When analyzing genomic data, we first need to align to the genome. There are a lot of possible choices in this, including BWA (medium choice), stampy (very accurate) and bowtie2 (very fast). Recently a new aligner came out, NextGenMap. It claims to be both faster and deal with divergent read data better than other methods. Continue reading
Tag Archives: DNA
Quick update: 96-well plate CTAB DNA extraction with fresh tissue
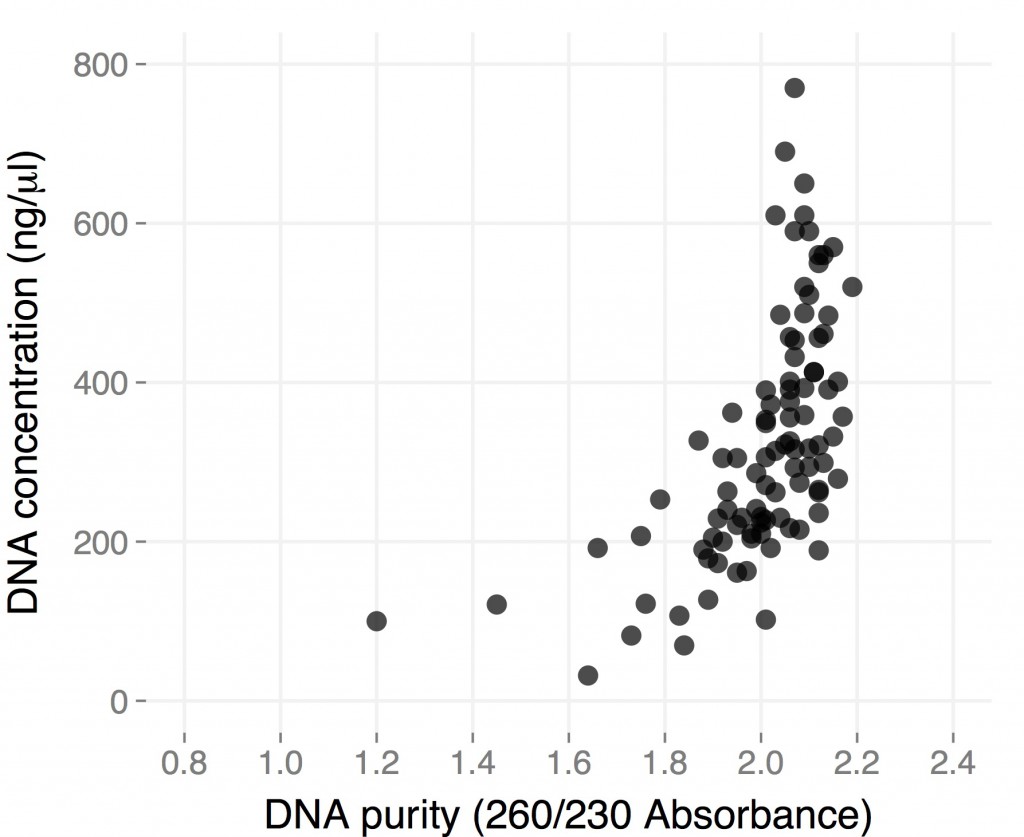
When I posted the protocol I have been using for CTAB DNA extraction in 96-well plates, I included results from a few plates I did starting from dried H. anomalus leaves I collected a couple of months earlier in Utah. While they showed that the method worked well enough when starting from “difficult” material, they were not exactly what you’d dream of when you decide to extract DNA, especially if you are starting instead from fresh material.
Here are the results from a plate of extraction I did starting from individual small (1.5-2 cm in length), young leaves from ~3 month-old H. anomalus plants. I collected the leaves directly in 96-well plates (I already put one metal bead in each well), put them on dry ice until I got to the lab (a couple of hours), left them overnight in the -80, and started extracting DNA the day after.
The final volume was 50 microliters, so total yield is for most samples between 10 and 30 micrograms of DNA. These are “real” DNA concentration measured by Qubit. Both average yield and purity are considerably higher than for dry tissue, and they are comparable to what you would get starting with frozen tissue using the single tube protocol (but you save a ton of time). Hope this gets you all more thrilled about 96-well plate DNA extractions 🙂
Paired End for Stacks and UNEAK
Both stacks and uneak are made for single end reads. If you have paired end data here is a little cheat that puts “fake” barcodes onto the mate pairs and prints them all out to one file. It also adds the corresponding fake quality scores.
perl GBS_fastq_RE-multiplexer_v1.pl BarcodeFile R1.fastq R2.fastq Re-barcoded.fastq
BarcodeFile should look like (same as for my demultiplexing script) spaces must be tabs:
sample1 ATCAC
sample2 TGCT
…
# note this could also look like this:
ATCAG ATCAG
TGCT TGCT
…
As it does not actually use the names (it just looks at the second column).
Here it is:
GBS_fastq_RE-multiplexer_v1
96-well plates CTAB DNA extraction
When I was working with Arabidopsis, 96-well CTAB DNA extraction was my best friend, and I spent many days extracting away tens of thousands of samples. Good times.
DNA extraction is much less pleasant in sunflower, but since I was reasonably happy with the results of single-tube 3% CTAB DNA extractions, I though I would try to scale it up to a 96-well plate format. Results of earlier attempts, with the participation of Brook and Cris, ranged from inconsistent to disastrous. Things all but improved when I tried again after coming back from Utah with a few hundreds dried samples. Since though the prospect of extracting them all one by one didn’t sound very attractive, I put some more effort into improving the protocol, and now it works quite nicely.
Plant DNAzol
Hi all,
Here’s another method for DNA extraction. The blog is stuffed to the gills with DNA extraction methods. The current standards are ‘Qiagen-like’ columnless – used to generate the DNA for the genome sequencing project and CTAB. I add this protocol because itis
easy and it is effective.
I extracted from Ha89 and harvested ~ 115 ng/uL from 20 cm tall plants. I also purposely ‘took it slow,’ letting tissue thaw after freezing to see PlantDNAzol’s efficacy. It’s efficient.
Quality is good
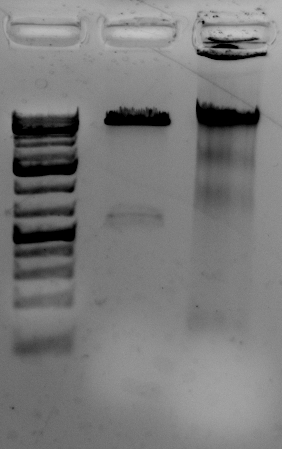
The gel to the left:
Right most lane contains Ha89 genomic DNA – 10 uL loaded of 70 at 115 ng/uL DNA – 260/280 was 1.78. 260/230 was ~1.00 (I suspect I could have added an additional ethanol wash to remove Guandine from the DNA mixture.
Is the DNA useful? Can downstream reactions proceed?
Yes. I digested the DNA with a methylation sensitive restriction enzyme, PstI and a methylation insensitive enzyme, EcoRV
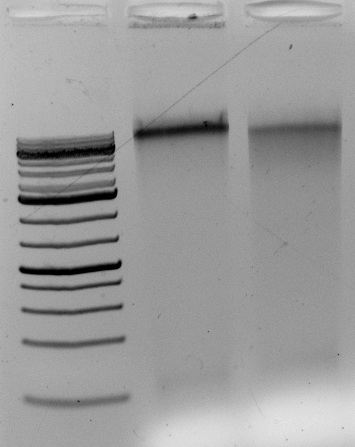
<–The gel to the left :
Leftmost lane Ladder
A vector digested with PstI,
Ha89 gDNA digested with PstI 240 minutes,
Ha89 gDNA digested with EcoRV 240 minutes.
The DNA digests. But does it contain contaminants that upset the enzymes over long incubations? Overnight?
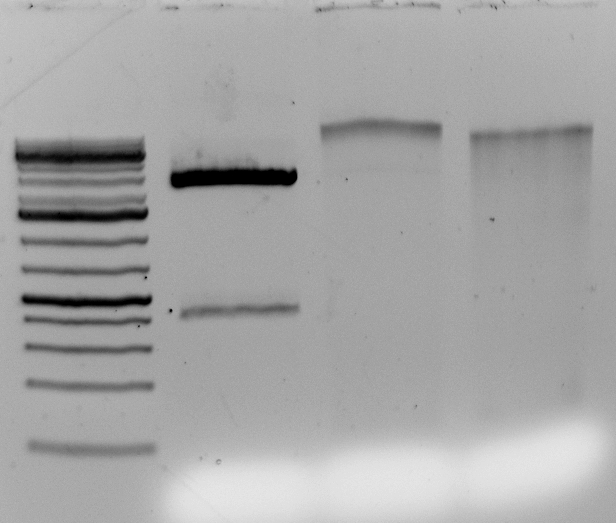
Gel above: From left- Ladder, unrelated vector digest,
Ha89 gDNA digested with PstI 22 hours
Ha89 gDNA digested with EcoRV 22 hours.
Protocol for DNAzol extraction (exactly as published by LifeTech but easier to follow – http://tools.lifetechnologies.com/content/sfs/manuals/10978.pdf:
Have these items on hand:
1 0.6 mL DNAzol per 100 mg sample
2. 0.3 mL chloroform per 100 mg sample
3. Timer
4. 100% ethanol (0.225 mL per sample),
5. 75% ethanol (0.3 mL per sample)
Handle all inversions carefully. When you see invert or shake handle your samples gently
1. Mix 100 mg ground tissue with 0.3 mL PlantDNAzol – 100 mg is max. Overdoing will hurt your yield.
2. Invert gently to aid in lysis and dispersion
3. Once completely dispersed incubate at RT, 5 min, shake periodically.
4. Add 0.3 mL chloroform and mix.
5. Once completely dispersed incubate at RT, 5 min, shake periodically.
6. Centrifuge at RT, 12 000 g (NOT rpm) 10 min
7. Harvest the supernatant. – you’ll see a phenol/chloroform styled triple layer. The middle layer will be pulpy containing your cellulosic debris and proteins. Don’t collect the middle layer. Less is more
8. Mix supernatant from 7 with 225 uL 100% ETOH.
9. Incubate at RT, 5 min
10. Centrifuge mixture 5000 g 4 min – get preparing for step 11
11. Make a PlantDNAzol – Ethanol mixture: For one sample mix 0.3 mL PlantDNAzol with 0.225 mL 100% ethanol.
12. Discard the supernatant from 10 and mix it with 0.3 mL of the mixture prepared in step number 11
13. Incubate as in step 9.
14. Spin as in step 10.
15. Pour off supernatant
16 Wash pellet with 75% ethanol – 0.3 mL – this step can be repeated if your 260/230 isn’t adequate. Guanidine absorbs strongly in 230 nm wavelength
EDIT: repeat step 16 for a total of 2 washes.
17. Spin as in step 10.
18. Remove supernatant – if your samples are green repeat step 16.
19. Resolubilize your DNA in TE or NaOH. Make sure to run your bead of TE over the wall of the tube to collect your DNA.
EDIT – Less is more as is usually the case with DNA extraction. I harvested from 38, 60, and 100 mg of tissue. – A sweet spot for tissue quantity is 45 to 60 mg for the given amount of Plant DNAzol
Qubit quantifiication: 45 mg of tissue yielded 264 ng/uL ug/mL. 60 mg, 305 ng/uL. 100 mg, 12.4 ng/uL
Tissue dessication with table salt
There is a new paper published by Elena Carrió and Josep A. Roselló online early in Molecular Ecology Resources that suggests salt dessication of leaves dehydrates and prevents decay at levels similar to that of silica gel, with similar PCR results.
Large-grain silica is probably still the best option, but this would come in really handy if you come across something interesting that you want to collect but don’t happen to have silica gel with you.
Here is the main figure (link to the paper below):

Thanks to Maggie Wagner from the TMO lab who found this paper and sent it around. Here is a link to the full article:
http://onlinelibrary.wiley.com/doi/10.1111/1755-0998.12170/full
DOI: 10.1111/1755-0998.12170
How my PhD was saved by DTT, or: how to get moderate quantities of clean, unsheared DNA from tricky tissues
I have been trying for months to find a high-throughput solution to DNA extraction from lyophilized (freeze-dried) leaf discs of Helianthus argophyllus, a species known for its intransigent polyphenolics and low DNA yields. And I appear to have found one, thanks to Horne et al. 2004 (here) and Dow Chemical!
Ph*cking phenolics!
Hi guys,
I have extracted sunflower DNA a dozen times now. I get one consistent problem a gelatinous film coprecipitating with my DNA (carbohydrates/polysaccharaides). I also get sharp smelling phenolic compounds. These occur regardless of DTT/2-BME addition.
I found a paper detailing the use of 2-butoxyethanol (2-BE) to remove strawberry polysaccharides and phenolics. The researcher’s idea is to perform a two-step precipitation:
Silica gel: God’s gift to botanists
Another Rieseberg lab member was asking me this week about preserving plant tissues on silica gel for later DNA extraction, and this got me to thinking about the general idea. I thought I would make a post about it, since it’s a useful technique, even in the genomic age. A lot of you already know all about this, so apologies for preaching to the choir.
I put an extensive post on my own Research Blog, but here is the gist of it:
Normalizing DNA with the Robot
Im not looking to volunteer myself as the robot expert, this is just one of the things you need if you are going to use it. These are the files needed to normalize a plate of DNA. One instructs it to take the volumes of DNA from one plate and put them into another and the other is to add the water. I can also offer a poor picture of the machine itself.
Freeze and squeeze DNA extraction from gel
Hi all,
Here’s a really easy DNA gel extraction:
1. Go about cutting out your bands of interest in the usual way.
2. Next cut the gel band(s) into tiny cubes.
3. Place tiny cubes in an eppie.
4. Put eppie in -20oC for 15 to 30 min. The idea is to make the band freeze into a mini ice cube.
5. Knock out all of the gel band/cubes onto a 10 (long) x 5 (wide) cm piece of parafilm.
6. Fold the parafilm in half along the length while keeping your gel cubes along the fold in the parafilm. You should have a piece of parafilm folded lengthwise with gel cubes sandwiched between two layers of plastic.
7. Crush/squeeze the liquid out of the tiny cubes. Liquid containing electrophoresis buffer and DNA will separate from the agarose.
8. Collect DNA/electrophoresis buffer mixture with pipet and clean it by ethanol precipitation.
EDIT: Borate in the electrophoresis buffer is a ligase inhibitor. Make sure to thoroughly wash your DNA with 70% EtOH, at least 2 times.
More CTAB!
Since I had as well troubles to get DNA extractions working with the protocols going around in the lab, I tried out a few published protocols (+ the regular CTAB extraction I used for Arabidopsis), picked the one that performed best in my hands and played around with it a bit.
Here’s the result: Sunflower CTAB DNA extraction protocol v1.2
Momma’s CTAB
Like a few other people in the lab, I’ve been struggling with DNA extractions of late. I’ve tried several methods, including the “columnless” method that a few people are using. But I was having pretty spotty luck. Then, during a bout of methodological soul-searching, I also tried a CTAB protocol that used to be my go-to method. It failed me a few weeks ago, but in a second trial last week, it delivered the goods: loads of very pure, high molecular weight DNA. It might not work for all plants, and there do appear to be situations in which it is not the best option, but if anyone wants to get back to down-home, from-scratch extraction, the way momma used to do it, then give this one a try:
Please note: the results below are for Streptanthus (the plant I’m studying; Brassicaceae); I’m trying it for some Asteraceae right now, and will update the post once I have results.
Momma’s DNA Isolation via CTAB
GBS protocol Version 2.0
Hi all,
Here is the long awaited updated GBS protocol.
PROTOCOL ->>>> GBSv2.0
There are three main changes from the previous protocol.
-After digestion and ligation, all the product is kept so more attempts can be made at the PCR.
-The PCR uses Phusion Taq, has longer extension times and one additional cycle with more primers/Taq.
-Size selection is done using AMPure beads instead of gel extraction.
DNA Gel excision protocol
Most gel excisions stink. Qiagen and Promega tend to claim 80%+ efficiencies that aren’t really achievable. Here I’m describing Jen Sheen’s SiO2 method for DNA purification from agarose: http://www.plantmethods.com/content/pdf/1746-4811-6-1.pdf
Making Illumina Whole Genome Shotgun Sequencing Libraries – (Dan E.)
I’ve been making whole genome shotgun sequencing libraries (for the purposes of this post: WGSS libraries) to sequence sunflower genomes on the Biodiversity Centre’s Illumina HiSeq. I haven’t been doing it for very long and its likely that my approach will change in the future as costs and products change but, as of early 2012, I’ve landed on a hybrid protocol based on kits from an outfit called Bioo Scientific. I use the Bioo Sci. adapter kit and their library prep kit up to the final PCR step at which point I switch to a PCR kit from another outfit called KAPA. I also use a KAPA kit to quant libraries with qPCR. In this post I give a little context then describe what I do to make WGSS libraries . . . Continue reading
DNA ploidy and genome size estimation using flow cytometry (Dan B.)
Flow cytometry (FCM) is – in principle – a relatively simple technique that allows one to measure properties of cells or organelles as they are intercepted (individually) by a high-intensity light source. In plant biology, it is often used to analyze extracts of intact nuclei, with the aim of determining DNA content and (indirectly) ploidy levels (this is the application I will focus on here). A good reference (including troubleshooting tips) can be found here.
Ampure XP beads – DNA clean up and size selection (Dan E.)
Ampure XP magnetic beads have become popular for cleaning DNA from reactions and for size selecting DNA. WGS sequencing library preparation protocols, and the GBS protocol, involve one or more bead clean up steps. If you make next gen sequencing libraries you will use Ampure XP beads. Following is some basic information about them and my thoughts about using them, mainly for library preparations . . . Continue reading
Zymo Research Genomic DNA Clean & Concentrator Columns (Dan E. and Kate)
We have some columns in the lab that are for cleaning and concentrating genomic DNA. They are made by an outfit called Zymo Research and should be easy to find, look for a bright yellow box.
This post describes a test I ran of these columns. Continue reading
Shearing DNA for WGS Sequencing- Bioruptor (Dan E.)
This post is about fragmenting, or shearing, genomic DNA to a particular size range using the Rieseberg lab’s Bioruptor sonicator.
Most of the current whole genome shotgun (WGS) library preparation protocols for NGS applications start with fragmented DNA. Generally speaking, this starting DNA should be a certain size and, for multiple samples, consistently that size. This objective turns out to be quite a tricky thing to accomplish with the Bioruptor. Given that WGS sequencing will probably continue to be popular in the lab, I am posting here what I have learned so far about taking whole genomic sunflower DNA and smashing it to the size range that I want using the Bioruptor. If I discover anything else in future library preps I’ll add it below. If anybody else has useful tips please comment.