Like a few other people in the lab, I’ve been struggling with DNA extractions of late. I’ve tried several methods, including the “columnless” method that a few people are using. But I was having pretty spotty luck. Then, during a bout of methodological soul-searching, I also tried a CTAB protocol that used to be my go-to method. It failed me a few weeks ago, but in a second trial last week, it delivered the goods: loads of very pure, high molecular weight DNA. It might not work for all plants, and there do appear to be situations in which it is not the best option, but if anyone wants to get back to down-home, from-scratch extraction, the way momma used to do it, then give this one a try:
Please note: the results below are for Streptanthus (the plant I’m studying; Brassicaceae); I’m trying it for some Asteraceae right now, and will update the post once I have results.
Momma’s DNA Isolation via CTAB
It’s pretty basic, and I think that might be the advantage that it has over other, fancier methods. Overall, I’ve found that the single most important factor in the success of this method is the amount of tissue used. Less is definitely more, as they say. For plants that are fleshy or succulent, use less tissue. If using dry tissue, never use more than 20 mg. I have recovered as much as 40 µg of DNA from 20 mg of dry tissue, so you can err on the side of less tissue and still get plenty of DNA.
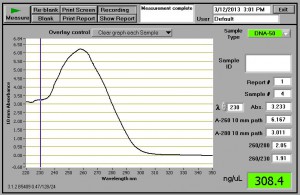
Here are some results that I obtained using 60 mg of fresh leaf tissue. This is with a 100 µL elution, so I could have as much as 30 micrograms of DNA from this extraction. The Nanodrop profile is for one of two samples; the other had about half this amount of DNA.
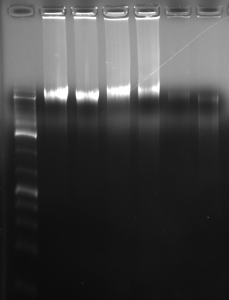
Here is the gel; the sample above is the fourth one from the left.
If you want to try it, let me know and I can lend reagents or run a few of yours myself.
Happy extracting!

Hello everyone,
Thanks for this Dylan. This protocol is an example of many standard CTAB based plant DNA extraction methods and it will be good to have it in our arsenal.
A question for Dylan: was the plant tissue in your demo extraction sunflower?
If it wasn’t it would be great to have a test run with some sunflower leaf. One of the several people currently in the lab doing DNA extraction might volunteer to try?
Note also that this method is very similar to our current CTAB protocol but without the CTAB precipitation step – i.e. taking the aqueous layer from the chloroform:isoamyl alcohol step and precipitating the DNA out of it with ethanol or isopropanol instead of with another CTAB buffer. It is that CTAB buffer precipitation step in our current protocol that is failing on us more frequently these days for some reason so it makes sense to eliminate it if we can. We just need to see if we get high quality DNA out of a simplified protocol like Dylan’s.
Also, this protocol does not have an RNase step in it. Just another difference between it and our current protocol. RNA often doesn’t matter but be aware that you will need to add and RNase step to Dylan’s protocol if you want to be sure to get rid of it.
Thanks again Dylan.
Dan.