When I was working with Arabidopsis, 96-well CTAB DNA extraction was my best friend, and I spent many days extracting away tens of thousands of samples. Good times.
DNA extraction is much less pleasant in sunflower, but since I was reasonably happy with the results of single-tube 3% CTAB DNA extractions, I though I would try to scale it up to a 96-well plate format. Results of earlier attempts, with the participation of Brook and Cris, ranged from inconsistent to disastrous. Things all but improved when I tried again after coming back from Utah with a few hundreds dried samples. Since though the prospect of extracting them all one by one didn’t sound very attractive, I put some more effort into improving the protocol, and now it works quite nicely.
The samples I have been using are dried leaves from H. anomalus plants, collected in September 2013. I don’t know if that’s common to all sunflowers, but dried H. anomalus samples seem to contain quite a lot of secondary metabolites – they have a distinctive smell and often produce small droplets of resin at the sites where they have been cut. Hopefully then, if this protocol works with them it should work for most other sunflower species, and give better results with frozen tissue.
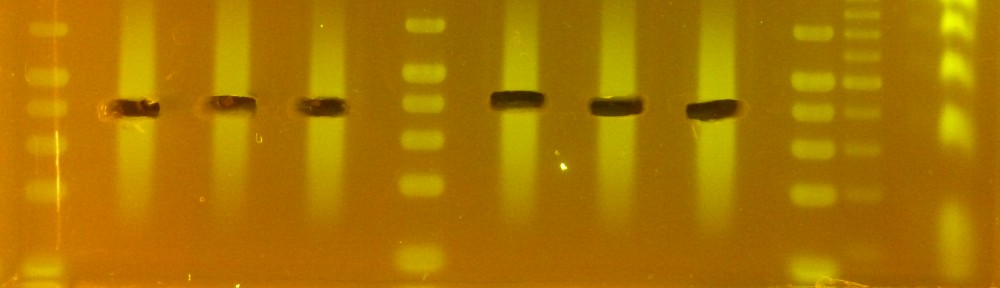
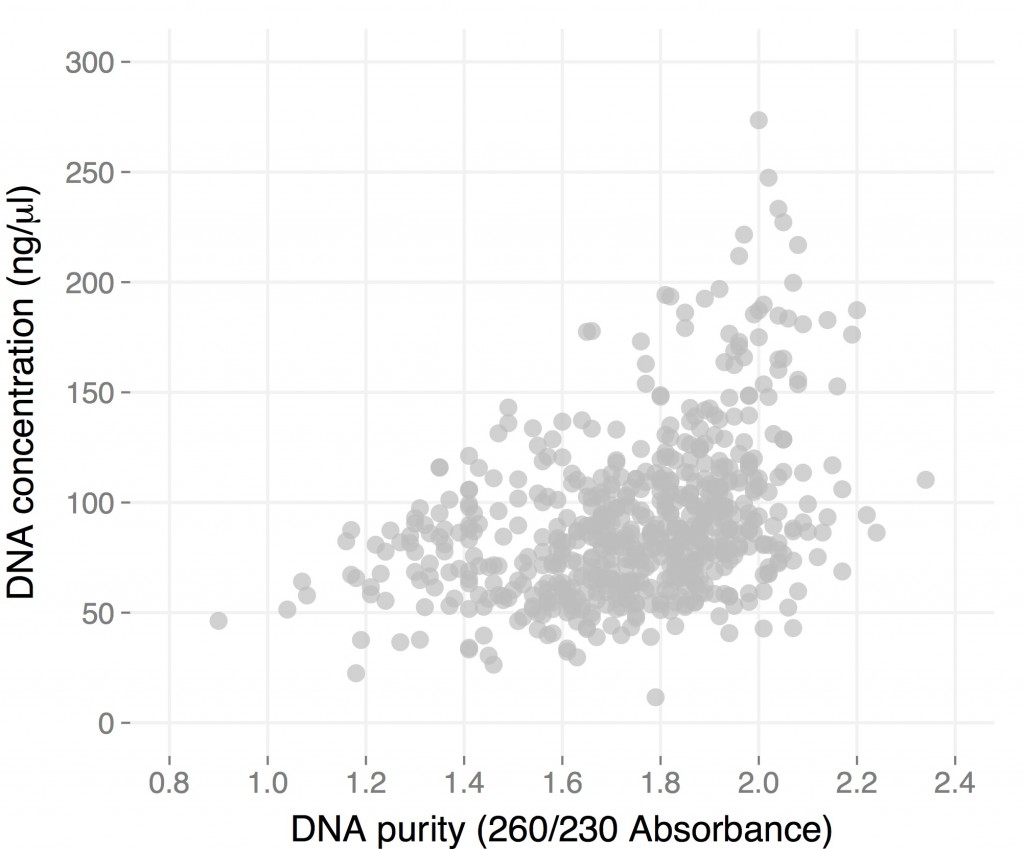
As I mentioned earlier, initially I obtained mostly nice brown pellets and very low-purity DNA (260/230 values in the 1.0-1.2 range). Shortening both the extraction and precipitation steps increased purity considerably with only a minor reduction in yield. These are the results for the about 600 samples I extracted:
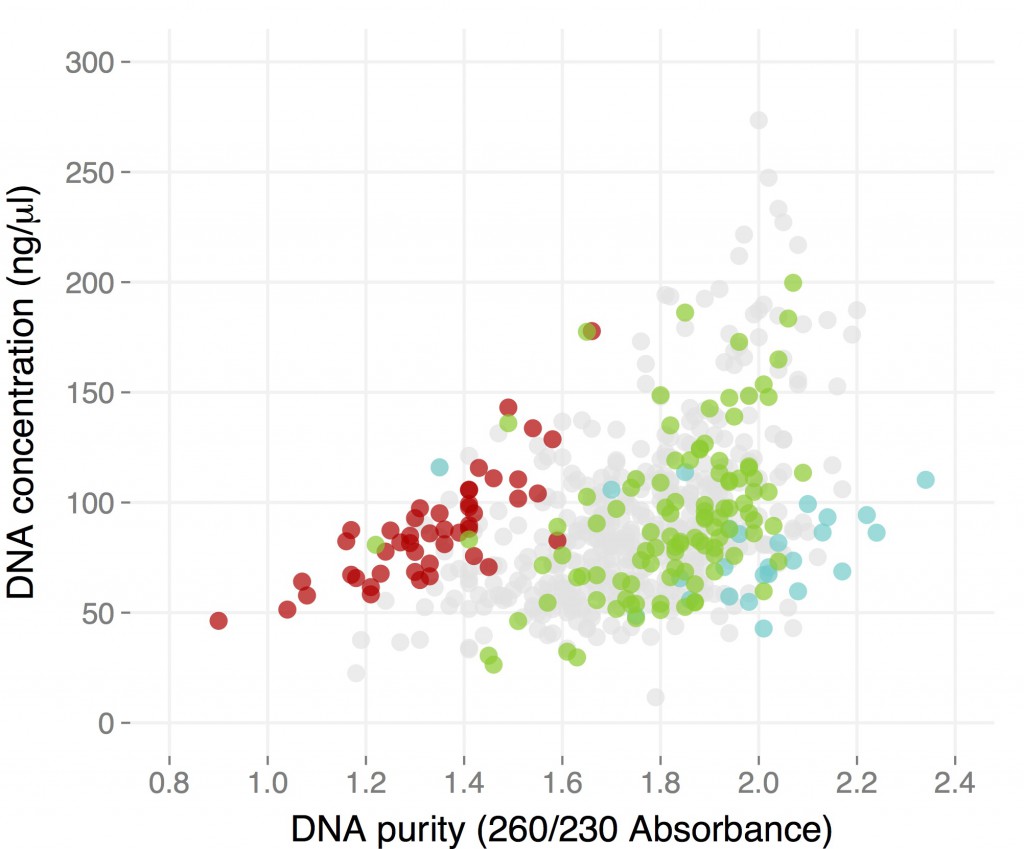
Considering that these are not the easiest of samples, I think this is quite good. These concentrations were obtained using a nanodrop, notoriously unreliable at this particular task. I re-measured some using the BR kit for the Qubit, and in this case the Nanodrop seem to over-exstimate DNA content by only a 30%. Since I resuspended the 100 microl of buffer, most samples have between four and 12 micrograms of DNA. 260/280 values were around 1.80 for all samples, and more than three quarters of them had 260/230 values of 1.60 or higher. These results are comparable to those I got by individually prepping the same samples. One factor affecting the results is the quality of the initial samples, which was quite variable: some came from large, happy plants, others from sick, semi-dry ones, and this reflects on the quality of the DNA. That is particular evident in the fact that some populations performed consistently well, and others consistently bad. See an example for three different populations in the plot below:
The protocol is not the fastest, but while the chloroform extraction in plate format requires a bit more concentration, the rest is just a lot of pipetting (make sure you have plenty of 300 microl tips before starting). The advantage is that it gives good quality DNA from moderately difficult samples. Also, if you plan to process several hundred samples, it is considerably cheaper than using a kit.
I added v1.3 of the protocol, with a few minor corrections