I’ve been making whole genome shotgun sequencing libraries (for the purposes of this post: WGSS libraries) to sequence sunflower genomes on the Biodiversity Centre’s Illumina HiSeq. I haven’t been doing it for very long and its likely that my approach will change in the future as costs and products change but, as of early 2012, I’ve landed on a hybrid protocol based on kits from an outfit called Bioo Scientific. I use the Bioo Sci. adapter kit and their library prep kit up to the final PCR step at which point I switch to a PCR kit from another outfit called KAPA. I also use a KAPA kit to quant libraries with qPCR. In this post I give a little context then describe what I do to make WGSS libraries . . .
Background
- If you have the time, its cheaper to make WGSS libraries yourself than to pay the premium to have it done for you. Doing it yourself also puts you in charge of all of the quality control, which you might appreciate.
- Illumina, to my mind, have screwed up. I’m just going to assume that they make a killing on the machines and the per-run consumables and they don’t care if people go elsewhere for the sequencing kits and reagents because, at least as of late 2011, the sequencing kits they offer are un-buyable. They cost a lot and they include only 12 barcoded adapters. I believe they now have two sets of 12 barcodes for a total of 24. Many of their potential clients (us!) want many more than 24 barcodes and, what’s more, we’d rather not pay the Illumina price when there are third party offerings that are substantially cheaper. If you want to check out the Illumina adapters and sequencing kits their brand, as of early 2012 is “Truseq”.
- One of the third party biotech companies is Bioo Scientific. As of early 2012, they are selling up to 48 6nt barcoded adapters (i.e. Illumina adapters with unique 6 nucleotide barcodes in them) and an alternative set of 96 8nt barcoded adapters as well as library prep kits that work and cost less than Illumina’s. I have used their adapters and library prep kits to make 61 WGSS libraries as of March 2012 and have no complaints, well, none that make me want to figure out what to do with just 24 very expensive Illumina adapters anyway.
- Obviously, the most economical approach to making WGSS libraries is to source your own adapters. (You could also work up your own end repair, adenylation and ligation reagents, but I’m not willing to go there). This would be very simple and considerably cheaper than paying a biotech company like Illumina or Bioo Scientific for them. The only thing you would be missing out on is the quality control that those companies provide. As of early 2012 I have not made any move toward this but the obvious thing to do would be to request the sequences for the Bioo Sci. 96 barcodes from their customer support and then order the oligos from IDT, then anneal them into double stranded adapters much like we do for the GBS adapters. I have the sequence information for the 48 6nt barcoded Bioo Sci. adapters already.
The Process – Overview
Read my post about Illumina adapters and follow the links therein to see a simple animation describing the basics of Illumina sequencing. In the same post there is a link to a nice pdf document made by someone from Tufts University which explains what the Illumina adapters do. Check it out if you don’t already know, its good.
My method for making WGSS libraries for the Biodiv. Illumina HiSeq machine developed in early 2012 is based on kits from Bioo Scientific and KAPA . . .
- Shear genomic DNA to the length that you want to sequence (Bioruptor or Covaris).
- End repair the sheared fragments, i.e. “blunt” them – fill in the single stranded ends created by the shearing (Bioo Sci. NEXTflex kit).
- Adenylate the fragments, i.e. add a single A overhang at both ends of each fragment (Bioo Sci. NEXTflex kit).
- Ligate Illumina adapters onto both ends of each fragment (Bioo Sci. NEXTflex kit and NEXTflex barcoded adapters). These adapters will attach the genomic DNA inserts to the flow cell that goes into the Illumina sequencer and provide the primer annealing sites for the amplification primers that are in play in both the cluster generation and sequencing steps of the Illumina process. They will also allow you to enrich your library by PCR in a later library prep step.
- ***Size select the fragments so that you have only fragments that fall into a size range that is suitable for the Illumina cluster generation process, usually somewhere between 200bp and 400bp.
- PCR enrich your size selected library using the Illumina sequencing primers (KAPA HiFi kit).
- Check the size of your library on the Bioanalyzer (a #$&^ing horror show).
- ******Quant the library using a qPCR kit (KAPA Library Quant kit).
- ******Dilute the library to the concentration required by whomever is running the Illumina sequencer you are using. As of early 2012, Ana wants WGSS libraries at 2nM for the Biodiversity Centre’s Illumina HiSeq.
- ******Once at the desired concentration you can pool barcoded libraries if you want.
*** The size selection can be done earlier. Bioo Scientific has two size selection options in their library prep protocol. You can do it with an agarose gel slice after the adapter ligation step or you can do it with Ampure XP beads after the end repair step and before the adenlyation and adapter ligation steps.
******You can probably get Ana to do steps 8-10. She may do a qPCR on your libraries even if you have already done it. If that is the case, you will be paying her for that and may be “wasting your time” doing it yourself. The benefit of doing it yourself, however, is simply your own quality control. The other factor here is the number of libraries you are pooling. If you are doing just a single library then you would do one qPCR and so would Ana. If you were doing 48 libraries and pooling them before handing that multiplex over to Ana you would do 48 qPCRs and Ana would do one. This means that if you gave Ana the individual libraries you would have to pay her for 47 more qPCRs. Talk to Ana and Loren about $$.
Kits, Beads and Chips
My library prep process revolves around kits from Bioo Scientific and KAPA. Note that there are alternatives, NEB is a popular supplier of products that are very similar to the Bioo Sci. and KAPA kits that I’ve been using so check them out and do some price comparisons. Also, there are several AMPure XP bead clean up steps and a Bioanalyzer run and at least one Qubit DNA quantification.
From Bioo Scientific:
- Barcoded Illumina sequencing adapters: NEXTflex DNA Barcodes, sold in sets of 6, 12, 24 or 48. Or their big 96 barcode set which I have not tried.
- Read my post about the above mentioned barcoded adapters and their sequences.
- End repair, adenylation and ligation enzymes and buffer mixes: NEXTflex DNA Modules.
From KAPA Biosystems (distributed by D-Mark Sciences):
- Library enrichment/amplification PCR kit: KAPA Library Amplification Kits.
- Library quantification qPCR kit: KAPA Library Quant Kits.
From Beckman Coulter:
From Agilent:
- Bioanalyzer kit: Agilent High Sensitivity DNA Kit.
From Invitrogen:
- Qubit DNA quant kit: Qubit dsDNA HS Assay.
Protocols
Here is the Bioo Scientific protocol for their NEXTflex library preparation kit. It is the basis of my method and I have annotated this pdf with a few pointers and tips.
Here is the Zymo Research Zymoclean Gel DNA Recovery protocol. I use these columns to purify DNA from agarose gel slices when I do the size selection with agarose.
Here is the KAPA HiFi Library Amplification Kit protocol. I use this to do the PCR step instead of the Bioo Sci. equivalent because its a bit cheaper.
Here is the KAPA Library Quantification qPCR protocol. I use this as the final QC step to get concentrations in nM so I can dilute and pool libraries.
————————————————————————————————
The Process – Tips
How much DNA do I need to start with?
Absolute: The Bioo Sci. protocol calls for 1ug of sheared DNA going in to the library prep. Where I have the luxury of samples with a lot of DNA I start with 3ug in the shearing step so that I can afford to lose quite a bit of it. This is, admittedly, a little excessive and if you are careful you might be able to routinely get away with less.
Note that this 3ug starting amount is my approach when using the Bioruptor to shear the DNA. You won’t need this much if you use the Covaris (see below).
If you don’t have 3ug or can’t afford to use that much of a DNA sample you can probably get away with less. Try to get as close as possible to 1ug of sheared DNA for the end repair step and, if you have substantially less, make a proportional reduction in the amount of the adapters in the ligation step.
Concentration: The Bioo Sci. protocol calls for 1ug of sheared DNA in 40ul or less for the end repair step. So, the minimum concentration of the 1ug DNA solution is 25ng/ul. Be aware of this when you design your Bioruptor shearing strategy. You will probably want to standardize the sample volume for the shearing step – just make sure your DNA concentration is at least 25ng/ul. My standard for shearing in the Bioruptor is 3ug in 75ul which is 60ng/ul, however, in an attempt to be less wasteful I did one batch of samples at 2ug in 75ul without considering the concentration, which is about 27ng/ul. Some of those samples were a little under the desired 1ug in 40ul (25ng/ul) after shearing.
If you have DNA samples that are too dilute, i.e. less than 30ng/ul or so, then concentrate them with columns (e.g. the Zymo Research DNA clean and concentrator columns) if you can afford to lose some of it or with the Adams lab speedy vac if you can’t. You can also recover samples that have become too dilute after shearing using the Adams lab speedy vac.
Qubit: Quant your DNA at least once with the Qubit. You want an accurate estimate so you can get as close as possible to the 1ug. You can quant your stock DNA before you take an aliquot for shearing and/or the sheared aliquot after you’ve finished shearing. I’ve done all of my samples both before and after so far. I have found that you generally don’t lose much in the shearing step. If you already have Nanodrop estimates for your DNA stocks and you are shearing a comfortable excess of DNA (like 3ug) you might not need the first Qubit estimate.
Shearing (about 1hr to several days depending on the number of samples and how careful/particular you are)
There are two options for this step, the Bioruptor in our lab and the Covaris at NAPS. I’ve used the Bioruptor exclusively so far. I may use the Covaris for special cases such as samples with very little DNA where all of it will be going into the library prep and I don’t want to risk wasting any of it on a misjudged Bioruptor blast. Bear in mind that the Bioruptor is almost free while the Covaris at NAPS is $10/sample (as of Feb 2012).
Read my post about shearing DNA for WGSS library preps using the Bioruptor.
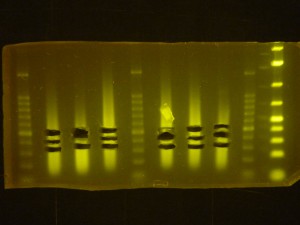
I aim to shear genomic DNA to 400bp for paired end sequencing.
Try to get DNA samples sheared such that the smear of DNA is centered at the desired size. Turn the exposure of your geldoc photo right up so you can clearly see where the main mass of DNA fragments is in terms of bp with reference to a ladder. The more DNA fragments of the right size you start with the more representative your library will be and the less PCR amplification you will need later.
Take a look at this image for an example of sheared DNA ready for the next step in the library prep process.
You can pause during and after the shearing, i.e. you can do bouts of Bioruptor sonication followed by agarose gel checks over the course of days and I’ve left the samples in the fridge for a couple of weeks before moving on to the next step.
AMPure XP Bead Clean Ups
There are several AMPure XP bead clean up steps coming up below. Read my post about using AMPure XP beads to clean and size select DNA molecules if you haven’t already. You are going to be doing it a lot
End Repair (about 90-120 mins)
This is the first enzymatic step in the process. The Bioo Sci. kit has an end repair buffer and enzyme that you add to your 1ug of DNA to fill in any single stranded ends on your sheared DNA molecules. Do it in a 96 well plate sealed with one of our silicone plate seals in one of our thermal cyclers. If your samples don’t fill the plate (probable) put them in the centre, or at least avoid the wells around the outside.
This step is followed by the first Ampure XP bead clean up. This bead clean up step takes considerably longer than the end repair reaction. Don’t be fooled by the Bioo Sci. pamphlet that says end repair takes 30 mins – that is the reaction time not the time it actually takes to get to a point where you can leave it.
You can pause after this step. Put your end repaired samples in the freezer. I’ve left samples like that for several weeks and nothing bad happened.
Adenylation (about 45 mins)
At this step you simply add a small volume of the Bioo Sci. Adenylation Mix to the final elute off the beads from the post end repair bead clean up. Mix it, then incubate in a thermal cycler again. There is no bead clean up after this reaction so it really only takes as long as it takes you to thaw the reagent, aliquot it into the wells, mix it and incubate it.
Don’t pause here, move on to the ligation step on the same day.
Adapter ligation (about 60-90 mins)
At this step you add a small volume of the Bioo Sci. barcoded adapter and ligation mix to the adenylated DNA from the previous step, mix it and incubate it for 15mins on a thermal cycler. This is where you give each of your samples its barcode. If you multiplex any of them on the sequencer, and even if you don’t, you will want to know which barcode each sample has. Be careful and write it down. There is a bead clean up step after the reaction which, again, takes a lot longer than the reaction itself.
If you started with appreciably less than 1ug of sheared DNA make a proportional reduction in the amount of adapters you add to the ligation reaction and make up the missing volume with the ligation buffer or water.
You can pause after the bead clean up. Put your samples in the freezer.
Size Selection (at least 90 mins and up to a couple of days)
There are two options here if you are using the Bioo Sci. NEXTflex kits: either you do an AMPure XP bead size selection back between the End Repair and Adenylation steps, i.e. you do a slightly more involved bead clean up step than the basic one you have to do between those two steps anyway, or you do an agarose gel size selection step here after the Adapter Ligation step.
So far (Feb 2012), I have done the agarose size selection for all but two WGSS libraries. Agarose works although it does take a lot longer than the bead method and there are tricks. My experience of the bead method has been a little disappointing. One of the libraries I made size selecting with beads had the desired 400bp mean fragment size but the other was around 1000bp. I suspect this was caused by pipetting error at the second bead step (too little bead mix would push the maximum size up). I redid the 1000bp one with beads again but was unable to get a size for it as the Bioanalyzer ate the remaining DNA HS chips.
Note that the Bioo Sci. protocol for the bead size selection is for 300-400bp fragments. If you want a different size you’ll have to do the agarose version or do some experiments with the bead mix concentration to move the size selection up or down.
Note also that the bead based size selection is done before the adapter ligation step so the size that is selected is the insert size. The agarose size selection, for some reason, is done after the adapter ligation step so the size selection is the insert size + the 121bp of the combined adapter lengths.
AMPure XP bead size selection: Be very careful about pipetting the bead mix into your samples. The difference in volume which moves the threshold size of DNA molecules that will precipitate on to the beads is very small – 2.5ul – and it is easy to transfer additional bead mix via the outside of tips.
If you haven’t already, read my post about AMPure XP beads, particularly the part about size selection.
How pipetting error will affect size selection:
- Too much bead mix at the first bead step -> move minimum size of the selection downward.
- Too little bead mix at the first bead step -> move the minimum size of the selection upward.
- Too much bead mix at the second bead step -> move the maximum size of the selection downward.
- Too little bead mix at the second bead step -> move the maximum size of the selection upward.
Combinations of the above might be particularly bad. If you did 2 and 3 you could end up with nothing – you could have overlapping size selections at the two bead steps that would result in the second bead step, where you discard the beads and the DNA bound to them, precipitating all of the DNA that you selected in the first bead step onto the beads. Errors 2 and 4 might have caused one of my first two bead size selected libraries to have a mean size of around 1000bp.
The bead method is much quicker than the agarose method especially if you have a lot of preps and can use the multichannel pipettes to speed things up. I would recommend being conservative with the multichannels however. I have had a frustrating time using them with bead cleanups. I would definitely use multichannels for the mixing steps and to aliquot and draw off ethanol, but If you want to minimize the transfer of beads at the elute steps you might find a single channel pipette works better.
Agarose gel size selection: There are a number of tricks to this.
- Read my post about agarose gel slices. I do the SYBR gold post-staining method and use the Sigma X-Tractor gel punches and the Zymo Research columns I’ve described in that post to do the WGSS library size selections.
- I run the libraries out with an empty lane between them. However, note that cross-contamination of libraries is actually not that much of a problem. If its not massive all you will have is minor contamination of one barcode identifiable library with another. You could probably even use the same gel punch for all of them, especially if you could rinse it in between punches.
- Note that I do one thing, that may be important, differently from the method specified in the Bioo Sci. protocol. I post stain the gel instead of adding SYBR Gold to the gel when I cast it. As you will read in below, there are difficulties with sizing adapter-ligated DNA in agarose. I was not aware of that issue when I started making WGSS libraries but what I was aware of is that SYBR-Gold in an agarose gel affects the migration of DNA by slowing it down, probably in a fragment size biased way. So, to minimize that problem I decided to post-stain my gels. Turns out that the Bioo Sci. people think that the SYBR Gold in the gel might help ameliorate the single stranded adapter end problem (see below) so, maybe I screwed up. If you want to try the agarose gel size selection method as specified in the Bioo Sci. protocol, please do, and then please tell me what happens with regard to your fragment sizes.
- Don’t forget that the adapters have been ligated on to your library of inserts. You need to add 121bp to the insert size that you are aiming for. If you want a 400bp insert, i.e. 400bp of sunflower DNA, you need to size select for 521bp DNA molecules.
- Even after adding the 121bp for the adapters you still need to compensate for differential migration of your DNA and the ladder. This is really tricky and we’d benefit from some more experimentation. The two DNA strands of the adapters beyond the 12 nucleotides proximal to the insert do not anneal to each other. These single stranded “tails” that are dangling off the ends of all of the adapter-ligated inserts interact with the agarose and slow the migration of the DNA through the gel. So, if you cut a band out at 521bp (400bp insert + 121bp of adapter) with reference to your entirely double stranded ladder, you will not recover a 521bp library. It will be smaller. Unfortunately the extent to which it will be smaller does not appear to be very consistent. If the exact size of your inserts is important to you and you have the time, some experimentation would be in order.
- If I get around to it I will make a graph or two and add them to this post but for now just be aware that for my first batch of 12 libraries I cut a slice from the gel at the 500bp ladder band and got mean fragment sizes (according to the Bioanalyzer) ranging from 398 to 425 (mean=410bp, std. dev.=9bp) which is 100bp smaller than it looked on agarose but not actually a disaster, or even a real problem, and at least it looked predictable. However, when I did another batch of 48 libraries and cut slices just above the 500bp ladder band I recovered mean fragment sizes ranging from 346bp to 580bp (mean=403bp, std. dev.=40bp) – yikes! Obviously, it looks like I did something different the second time. I don’t know what. Reassuringly, the average mean fragment size is, again, about 100bp smaller than the apparent size on a post-stained agarose gel but that variation is not good.
- If you do the agarose gel slice size selection it is a very good idea to take more than one gel slice. This will provide you with alternative insert sizes and a second chance should anything go wrong later. I would recommend taking a gel slice a bit bigger than desired insert size + 121bp of adapter + 100bp to compensate for slow migration, so ~620-650bp for a ~400bp insert, and another slice below that. To be really careful you could also take a third slice just below the second one.
- I use the Zymo Research gel DNA recovery columns to get the size selected library out of the gel slice. They work well. If I’ve got multiple gel slices per sample I only purify two of them. Additional slices I store as gel slices in the freezer.
- Use 10mM Tris to elute off the Zymo columns.
You can store the size selected libraries in the freezer.
PCR (minimum of a couple of hours – there is a bead clean up step)
After the size selection step, or ligation step if you did the Ampure bead size selection, you need to PCR enrich your library. The good thing about this step is that you get multiple goes at it if necessary. Doing second or third PCRs is costly, however, so don’t get carried away.
Note that the PCR step achieves a few good things:
- You get more of your library. Enough to sequence at least once.
- You enrich specifically for fragments that have the Illumina adapters on both ends. This means that the amplification of your library is actually an enrichment for the molecules you want.
- A successful PCR tells you that you have a “good” library, i.e. that you have molecules with the Illumina adapters ligated to both ends. A failed PCR is an indication that something has gone wrong with earlier steps in the library prep procedure, the ligation step for example. Note that a failed or low-yield PCR might be due to insufficient amplification cycles as well (see below).
Note that this PCR may also do one bad thing:
- PCR is generally sequence biased. If you throw genomic DNA into a PCR like this you probably won’t amplify all sequence equally. The KAPA HiFi taq is supposedly less sequence biased than other taqs but, still, this PCR will probably bias your library. To minimize this bias you want to do as few amplification cycles as possible.
I use a PCR mix from KAPA called “HiFi” which contains everything except the primers and the template. I haven’t tweaked anything except the template volume. The obvious tweak to try would be halving the reaction size to save money, although this might be difficult if you want to keep the template volume up in an effort to improve yield without adding amplification cycles (see below). I might try a smaller reaction volume the next time I make libraries.
The objective in the PCR enrichment is to get your library concentration up above the concentration required for submission to the sequencer. As of early 2012 Ana wants libraries at 2nM, so that is the bar you’ve got to get over. Its nice to have an excess of a library, however, so you can re-sequence it if you want to.
You also want to enrich your library with the fewest PCR amplification cycles as possible to keep any PCR bias down.
Most protocols say to do somewhere between 10 and 15 cycles.
The only parameter you can tweak, apart from the number of cycles, is the volume of your size-selected template DNA that you start with in the reaction. I maximise this in an effort to do one or two fewer amplification cycles.
So, the KAPA HiFi protocol specifies a 50ul reaction with 25ul of HiFi mix which gives you 25ul to play with for the primers and the template. For my most recent batch of 48 library preps in Feb 2012 I had my primers at 2uM (which was a bit dumb, I should have made them up more concentrated, say 10uM) which, in my world, is a 10X concentration of a PCR primer. So, I had to put 5ul of each primer into my 50ul reactions leaving me with 15ul of remaining volume for the template DNA – so, I put 15ul of my agarose gel slice size-selected libraries into the 50ul PCRs.
For my most recent batch of 48 library preps mentioned above I did an 11 cycle PCR then quanted them with the Qubit. I found that 9 of them quanted at less than 2ng/ul and I did a second PCR for these with 15 cycles. Note that this 2ng/ul threshold is almost entirely arbitrary (see below)! The 15 cycle PCR brought all but one of the 9 duds from the first PCR up to way over my minimum threshold (I probably could have done a 12 or 13 cycle PCR). The one library that failed to amplify twice was presumably an outright dud – proabably a ligation failure – and I redid that one from the beginning.
Library QC (at least a day and up to several – depends on the number of libraries)
As noted above, you might be best served handing you libraries over to Ana for QC and pooling/dilution. Even if you do give them to Ana you should do the QubitHS quant described below because that will tell you if the PCR step has worked or whether you should try the PCR again.
There are three QC steps that you can do and I do all of them:
- A QubitHS quant.
- A Bioanalyzer run.
- A qPCR.
The QubitHS assay and the qPCR are both for quantification. The qPCR is the ultimate in library quantification but takes time and costs more than the Qubit. I do both because the Qubit can be done quickly and tells me whether I need to do the PCR enrichment again while the qPCR probably gives me a more accurate quant for pooling and dilution.
The Bioanalyzer run also provides a quantification but you really do it for the sizing and to see if you’ve got contaminating adapter or primer dimers or multimers.
Qubit HS (depends on the number of libraries – up to an hour)
I do this straight after the PCR enrichment to see if I need to do it again, usually with more amplification cycles. My threshold for this call is 2ng/ul which is not quite entirely arbitrary. I have QubitHS and qPCR quants for a number of libraries now and every one of them that quanted at >2ng/ul with the Qubit was above 5nM according to the qPCR. Note that, as of early 2012, you need your libraries to be at least 2nM to give them to Ana for sequencing.
Bioanalyzer (you can only run 11 libraries per 45 min run and there will probably be re-runs – so 1hr to several days)
This sucks. Read Kathryn’s post about this monstrosity and ask around to see if anybody has used it recently and has any tips.
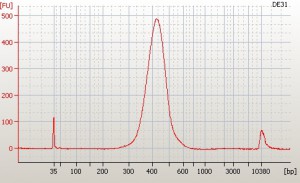
I run 1ul of my completed libraries (i.e. have passed my post-PCR Qubit test above) on a DNA High Sensitivity Bioanalyzer chip and do my best to make it work. When it works its great – you get to see if your library is a nice single peak and what the mean size of it is and whether you’ve managed to get rid of all the adapter and primer products with the Ampure bead clean ups. This is what it should look like:
Those two peaks on the left and right of the library peak are markers not contaminants.
If you want to take the qPCR seriously you need to know the mean size of your library of DNA molecules.
To get the size, and quant, information out of the Bioanalyzer you need to do a “Region Table” in the Bioanalyzer analysis software. Set the minimum and maximum bounds for your region immediately around the library peak.
Get a report out of the Bioanalyzer after you’ve done all of the region table analyses. Its a good idea to put the library identifiers into the Bioanalyzer software so you can keep track of them. You want a “. . . _Results.csv” file and probably also the .jpg files for each sample and the “gel”.
qPCR (the first time you do it will take longer – a few hours to one day)
I use the KAPA library quantification qPCR kit for Illumina libraries for this step. It is simply a PCR master mix without the primers and six concentration standards that you include in your qPCR as references. You can use generic qPCR kits of course. The KAPA kit is just easier and it has worked very well for me so far.
Yes, I do do it in triplicate – that means each standard and each library is done three times.
You need to dilute your libraries for the qPCR. If they are concentrated enough to have passed my 2ng/ul Qubit threshold, i.e. probably at least 5nM and possibly orders of magnitude more concentrated than that, they will be way too concentrated for the qPCR to resolve.
I’ve only done this twice, as of March 2012. The first time I dilute libraries 1/1000 and 1/2000 but these were too concentrated, i.e. the exponential amplification took off before the most concentrated of the standards. Extrapolating outside of the standards is not crazy but its not ideal. I redid those libraries with a 1/8000 dilution and they were still at the more concentrated end of the spectrum of standards. The second batch of libraries I qPCR’d I diluted 1/4000. Again a bit concentrated but within the range of the standards.
Dilute an aliqot of your libraries in 10mM Tris, 0,05% TWEEN20.
To get to the dilution you ultimately want you will probably have to make serial dilutions – like 1 in 100 then 1 in 20 to get to 1/4000. For example, I took 2ul of my libraries and added that to 198ul of dilution buffer = 1/200. Then I took 5ul of that 1/200 dilution and added it to 195ul of dilution buffer = 1/40 dilution for a total dilution of 1/4000.
Be very careful about pipetting the correct volumes for the dilutions and the qPCR itself. Pipetting error will be the most serious, semi-preventable, source of error in the qPCR quanting.
You do the qPCRs in triplicate to control for pipetting error when setting up the actual qPCR, most importantly when you aliquot your template DNA (diluted library) into the PCR plate. Obviously, this does not control for pipetting error at the dilution steps. For this reason, among others, you cannot expect your qPCR quants to be perfect.
Turn the Biorad I-cycler qPCR machine on – both the thermal cycler and the lid where the camera is (2 on switches) while you make up the reaction – the thing needs to heat up.
Run the PCR described in the KAPA protocol.
Select “Persistent well-factors”.
Ask around if you want help setting up the qPCR in the Biorad software.
The machine will automatically calculate concentrations and the mean of the triplicate reactions. Its simple to get a report in a text format that you, unfortunately, can’t just open in Excel. I copy and paste the columns of data that I want into Excel.
You need to do a little arithmetic to go from the concentrations the machine calculated, which are the concentrations of template DNA in the actual reaction relative to the standards, to the concentration of your libraries. You need to account for the dilution and for the difference in bp size between your libraries and the standards.
Its easy – Take the mean concentration of the templates from the three qPCRs in pM, as calculated by the qPCR software, and multiply it by 452 (bp size of the KAPA concentration standards) divided by the mean size of your library (from the Bioanalyzer run), and multiply by the dilution factor (e.g. 4000). That is the concentration of your library in pM. Divide that by 1000 for nM.
For example, one of mine:
- Mean template concentration from 3 qPCRs = 12.8pM
- Mean size of library = 403bp
- Size of KAPA kit concentration standards = 454bp
- Dilution factor: library stock to template used in qPCR = 4000
- Concentration of library = 12.8*452/403*4000 = 57425pM = 57.4nM
Dilution (up to a couple of hours depends on the number of libraries)
Dilute your libraries to the concentration required by the sequencing facility. As of March 2012 Ana wants libraries at 2nM for the in house Illumina sequencer.
If you are pooling barcoded libraries to be run in a single lane, the easiest way to do it at a particular concentration, like 2nM, is to dilute the individual libraries to the desired overall final concentration, 2nM for example, then pool equal volumes of each diluted library.
You can use 10mM Tris or 10mM Tris, 0.05% TWEEN20 as a dilution buffer.
Based on my experience so far you will be diluting your libraries a lot but there will be a lot of variation among them. If you are doing more than one library the easiest way to do it is to calculate the volume of dilution buffer that you need to add to a set volume of each library stock to achieve the correct concentration. This way you will take the same small volume of each library, and not have to pipette ridiculously small volumes.
The way I do it:
- Do the dilution calculations in Excel and print it out. Work out the volume of dilution buffer that will be needed to add to a 3ul volume of each library. For example, lets say I have a library at 30nM:
ul to add to 3ul of 30nM library to get 2nM = (3*30/2)-3
I always round that calculation up or down to an integer in Excel.
- Aliquot the volumes of dilution buffer to suitably sized wells or tubes, I use 1.6ml tubes.
- Note that you can change the 3ul volume of the library stocks to bring the total volume of the dilution up or down. Don’t make it too small, you want to pipette it accurately.
- Add the 3ul of library stock and mix. Done.
That is it. If you are sequencing on the Biodiv. Illumina HiSeq, find Ana and get the submission form, fill it out and give it and your tube(s) to her.
Get a beer.
Dan E. March 2012.