Filtering the variants is a critical step in the pipeline, as most of the SNP callers are very inclusive by default. Below are the criteria I have used to filter SNPs:
Author Archives: Kay
SNP calling V – SNP and genotype calling
The next step in the pipeline after the alignments are conducted and adjusted is to call the SNPs and genotypes. There are many programs that will do this such as the ML script that many people in the Rieseberg lab use, freeBayes, mpileup and bcftools, as well as the GATK UnifiedGenotyper. To bench mark the alignment steps in these SNP calling blog posts I used mpileup and bcftools. For example:
SNP calling IV – Base quality score recalibration
The best practices protocol from Broad highly recommends base quality recalibration (http://gatkforums.broadinstitute.org/discussion/44/base-quality-score-recalibration-bqsr). Apparently, the base quality scores off the machine are not very accurate and can provide false confidence in the base calls and consequently influence SNP calling. This tool will adjust average quality scores and also adjust the scores depending on the machine cycle and sequence context (i.e. start versus end of read, type of dinucleotide repeat). After recalibration, the quality score is closer to its actual probability of mismatching the reference genome.
To run this you need a database of known SNPs so the tool can assess the error rate at other sites based on the actual sequence data. Unlike humans, which this tool was designed for, most species do not have comprehensive SNP databases. However, such a table can be created by identifying SNPs in a preliminary run without the base quality score recalibration. I included any SNP that had a Qual score of 20 in my SNP vcf file.
You can see below that it had a large effect on the number of SNPs that I called. With the BAQ filter off 263,360 SNPS overlapped between the methods. 14,591 were unique to the recalibrated alignments and 131,354 were unique to the alignments without base quality recalibration.
SNP calling III – The indel problem
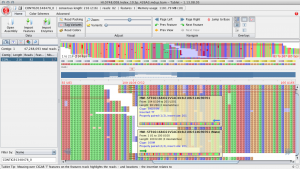
SNPs can often be falsely called around indels. In particular, if indels are near the start or end of a read the read is often incorrectly aligned. Here is an example of how things can go wrong from a spruce alignment. Below, at site 109 there is a TT insertion in the reads relative to the reference and there is no true SNP at this site. However, this individual was called as a het (C/T) because of the reads that end in the middle of the insertion. For these reads the penalty for a single mismatch to the reference is less than introducing a gap.
SNP calling II – Creating a reference for GATK and Picard
Using GATK and Picard was difficult because we had to keep modifying our reference (and therefore redo the alignments each time) to use these programs efficiently. Below I list the things we had to do to the reference to get them to work:
SNP calling I – alignment programs and PCR duplicates
I have tested various SNP calling methods using exome re-sequencing data from 12 interior spruce samples. I tried Bowtie2, BWA (mem), Picard (mark duplicates) and GATK for indel realignment and base quality recalibration. For SNP calling I used mpileup with and without BAQ as well as the Unified Genotyper from GATK. For an interesting and informative workshop outlining the Broad best practices SNP calling pipeline check out these youtube videos (http://www.youtube.com/watch?v=1m0ZiEvzDKI&list=PLlMMtlgw6qNgNKNv5V9qmjAxbkHAZS1Mf). My results are in a series of blog posts and I hope you find them useful. Please let me know if you have any suggestions for SNP calling. We only want to do the alignments and SNP calling once for the entire set of samples, because it is going to take a long time!
Blast2GO
This describes how you can run blast2go on a server using b2gpipe and a local database. This makes blast2go a viable option for annotating large fasta files. Otherwise it is much too slow. The database is currently set up on an AdapTree server. This took a while for me to troubleshoot, so you could run into different problems, but you will hopefully avoid some of the issues I ran into. The b2g Google group is good for troubleshooting. You can find many of these instructions at http://www.blast2go.com/b2glaunch/resources/35-localb2gdb