I’ve done a small analysis on my GBS data and posted it on my blog: http://www.proseedwithscience.com/?p=816
Edit: This is mostly just a quick look at the amount of missing data in the data and some potential explanations of where it might be from.
I’ve done a small analysis on my GBS data and posted it on my blog: http://www.proseedwithscience.com/?p=816
Edit: This is mostly just a quick look at the amount of missing data in the data and some potential explanations of where it might be from.
This describes how you can run blast2go on a server using b2gpipe and a local database. This makes blast2go a viable option for annotating large fasta files. Otherwise it is much too slow. The database is currently set up on an AdapTree server. This took a while for me to troubleshoot, so you could run into different problems, but you will hopefully avoid some of the issues I ran into. The b2g Google group is good for troubleshooting. You can find many of these instructions at http://www.blast2go.com/b2glaunch/resources/35-localb2gdb
Hello All,
Many of us have been annoyed by the restricted file types that WordPress allows to be uploaded to RLR. It’s especially annoying because all WordPress is doing when it permits or denies an upload is checking the file extension against a list of allowable extensions. {Even the most malicious code could be uploaded to our blog as long as it had a .txt file extension. Whether that code could then be made to execute, however, is far beyond my web-programming grasp – WordPress would treat it as plain text so it may be impossible.}
We’ve been sharing code via RLR by sidestepping the file extension rules and uploading scripts as .txt text files or by compressing files into zip archives or just putting the code itself into posts. Admittedly these were simple solutions, but now it’s even simpler – I just added some of the relevant file extensions to the list that RLR will allow for upload.
I added: “.pl”, “.py”, “.sh”, “.R”, “.r” and “.kml”.
Any file with one of those extensions will upload as plain text, i.e. WordPress will treat it as a text file.
If I’ve omitted something useful let me know.
Please remember that code can simply be copied into the body of a post and that will often be the best way to share it. But, in addition to that presentation, and especially for long scripts, you can now upload the script with its file extension to the RLR media library and put a link to it in your helpful post explaining what it does.
Dan.
Some SNP table to useful table conversion scripts are here: FormattingScripts_v0.4
Readme.txt explains usage, makes fasta, bayescan, structure files as well as converting to digits for R.
Let me know if you find any of this useful or broken.
Greg
Edit: updated small fix to structure formatter
I couldn’t believe how expensive the software was for writing barcodes, so I wrote a short program in R to do it for FREE. And, frankly it should be faster and easier if you already have your labels in an Excel file. You don’t really need to understand the program or even R functions to use it, as long as you know how to run an R program.
Setup and Overview:
[UPDATED (see notes below)] – R-code. Start with this (Note I could not upload a .R file, so this is .txt but still an R program).
Input – barcodes128.csv – You need this file to run the program. Save it in your working directory (see comments in R code for how to set this). AND labels.csv – This is a sample file showing the format for your labels. Even though it’s a .csv, it is a single column with each label as a separate row, so there are no actual commas
Output – BarcodesOut.pdf – A sample output: a pdf file for the 0.5″x1.75″ Worth Poly Label WP0517 (Polyester Label Stock), currently in the lab
That’s really all you need to know, everything that follows is extraneous info. If you have any problems, check out the Detailed Instructions, Troubleshooting Tips, or add a comment below. Continue reading
I’ve installed the latest version of Ubuntu (12.04) on the old PC lab computer:
-Username, computer name and password are written on the computer itself, if needed.
-I’ve also installed on it a few of my favorite programs (LibreOffice, Inkscape, Gimp, R, Chrome).
-It boots in about 35 seconds, not bad for an “old piece of junk”!
Feel free to use it!
seb
Snowhite is a tool for cleaning 454 and illumina reads. There are quite a few gotchas that will take you half a day to debug. This wiki has a lot of good tips.
Snowhite invokes other bioinformatics programs, one of them being TagDust. If you get a segfault error from TagDust, it may be because you are searching for contaminant sequences larger than TagDust can handle. TagDust can only handle maximum 1000 characters per line in the contaminant fasta file and maximum 1000 base contaminant sequence lengths.
A segfault (or segmentation fault) happens when a program accesses the wrong piece of memory. After TagDust hits the 1000 line character/sequence base limit, TagDust keeps trying to access memory past the 1000 memory slots it has allocated. It may try to access non-existent memory locations or off-limits memory locations. You need to edit the TagDust source code so it allocates enough memory for the sequences and does not wander into bad memory locations.
char line[MAX_LINE];
MAX_LINE to a number larger than the number of characters in the longest line in your contaminant fasta file. You probably can skip this step if you are using the NCBI UniVec.fasta files, since the default of 1000 is enough.
char tmp_seq[MAX_LINE];
MAX_LINE to a number larger than the number of bases in the longest contaminant sequence in your contaminant fasta file. I tried 1000000 with a recent NCBI UniVec.fasta file and it worked for me. make clean in the same directory as the Makefile make clean in the same directory as the Makefilerelocation truncated to fit: R_X86_64_PC32 against symbol” errors during linkage. This occurs when the compiler is unable to allocate enough space for the program’s statically allocated objects. Edit the Makefile so thatCC = gcc
becomes
CC = gcc -mcmodel=medium
Last year I worked on a project to see if any of the domestication outlier genes were found with previously mapped QTLs. The project ultimately fell flat when new data showed that the outlier I was working on wasn’t an outlier, but I did compile a large table of sunflower QTLs which may be useful. The table has 369 mapped QTLs.
I’ve shared this with a couple of people, but I’m posting it here on a google doc for everyone to use. Here is the link: https://docs.google.com/spreadsheet/ccc?key=0AgfXIvTZMEqPdHdJWTk3UVlVa3dkdGFTak9ySlUtNkE
A couple notes:
-It was compiled about a year ago, so it may be out of date. Also, although I tried to include every applicable study, I may have missed some. If you do find a study that I missed, I encourage you to add it to the table.
-It is only from annuus crosses, and a majority are domestics
-The position values are in cM
Anyway, read and enjoy. Change it if you find errors or new papers!
There are a few publicly available data sets that are useful for looking at the abiotic environments of specific locations.
I’ve generated three training data sets, which will save you around 5 days if you decide to run Jaatha, a molecular demography program. It uses the joint site frequency spectrum of two populations to model various aspects of population history (split time, population size and growth, migration). Here’s the paper: Naduvilezhath et al 2011.
1. Using the default model, with the following maxima: tmax=20, mmax=5, qmax=10.
2. Alternative maxima: tmax=5, mmax=20, qmax=20.
3. Alternative maxima: tmax=5, mmax=20, qmax=5.
They can’t be uploaded because they’re compressed R data structures, but let me know if you’d like to give them a whirl.
Occasionally I find myself reading a news item or a paper that mentions a particular sequencing platform and scratching my head to remember of what exactly that particular platform is capable. If you ever find yourself in that same boat, the Molecular Ecologist has a very handy and often-updated guide here.
I’ve been writing posts about the various steps involved in making WGS sequencing libraries for the Biodiversity Centre’s Illumina HiSeq machine and I was tempted to try to explain what the adapters are for. Then I had the bright idea of seeing if somebody else had already done it. Somebody has. Continue reading
As of March 2012 we are using the Bioo Scientific NEXTflex barcoded adapters for WGS sequencing libraries made by ourselves, (well me so far). The set we are currently using comprises 48 barcodes, so we can multiplex up to a 48-plex in one lane on the Illumina HiSeq sequencer.
Below are the sequences of the Illumina adapters and the 48 barcodes we are currently using. Continue reading
RSEM is a relatively new bioinformatics tool that has been developed in conjunction with Trinity for the analysis of RNAseq data. RSEM can be used to estimate expression levels for both genes and different isoforms of genes, and is quite quick and easy to use, with an excellent google group for help (“RSEM users”). All it requires is an RNAseq dataset (either fasta or fq format) and a reference transcriptome that it can be aligned to.
GNU Parallel makes it easy to take advantage of multiple processors at once. I really enjoy using it to parallelize Bash scripts. For example, here’s a pipeline that will generate a reference-based transcriptome from Illumina-sequenced ESTs: . . . Continue reading
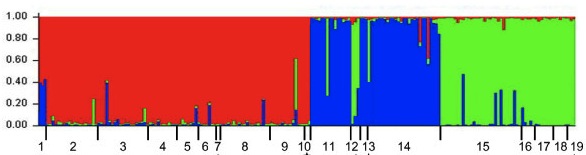 I suspect there are probably several homemade versions of this kind of script kicking around, but here is a perl script I’ve written for turning your SNP table into a STRUCTURE input file. To use it, you should change the .txt to a .pl after downloading the script. More on STRUCTURE input files (and so much more!) is in the documentation here.
I suspect there are probably several homemade versions of this kind of script kicking around, but here is a perl script I’ve written for turning your SNP table into a STRUCTURE input file. To use it, you should change the .txt to a .pl after downloading the script. More on STRUCTURE input files (and so much more!) is in the documentation here.
Recently BWA (an alignment program) suddenly started giving a strange error message, indicating that a reference file ending in *.nt.ann was missing. This file type was unfamiliar to me, with good reason: it’s a colourspace reference file, which shouldn’t be generated when we index the fasta-based references we’re using (at least, I don’t know of anyone in our lab using SOLID data as a reference). DO NOT rebuild the reference with the -c (colourspace) flag, as you might see suggested on the web, because we don’t know what effect that might have on our alignments. DO rebuild it with the usual settings.
Hi guys,
Following up on Nolan’s bioinfo workshop last week, here is a copy of
Alistair’s cheat sheet from the workshop he gave last year. I have
found it quite useful.
cheers,
seb
Bioportal is a free computing resource that provides several applications in our area. I’ve been running STRUCTURE on both the “low priority” and normal queues and it’s been fantastic (unlike Westgrid, who haven’t even responded to my application). For those of you who are struggling to find room on the cluster, it might be useful to you too. Much as I’d like to keep it to myself and exploit the hell out of it, here’s the address:
https://www.bioportal.uio.no/