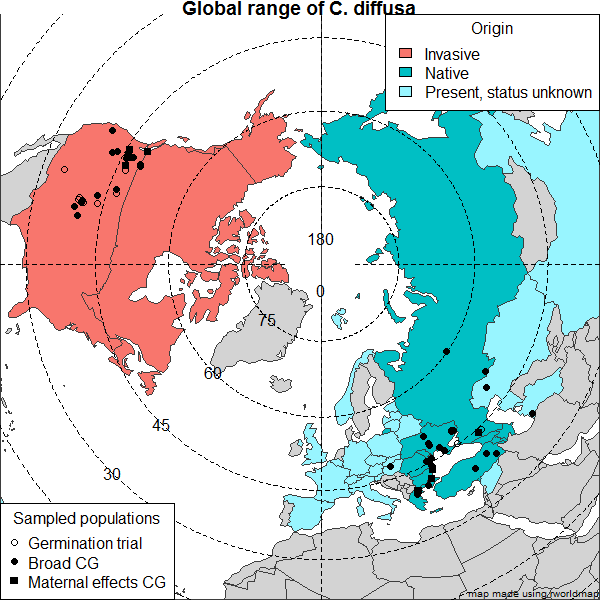
Helianthus germination
Everyone in the lab seems to have their own brand of germination suited to their seed collections. If you have fresh seed Nolan’s protocol easy sunflower germination is a great place to start and germination should occur within a week (I have found this works with fresh H. paradoxus seed).
Here are is an updated version of Nolan’s germination protocol. I am working with some older Helianthus seed that takes several weeks to germinate. I have received useful advice from several lab members who are referenced within. I haven’t yet found a way to speed germination of old seed, but the emphasis here is on how to keep conditions sterile to prevent fungal infection until germination occurs. Patience is required.
Feel free to correct or add your brand of germination to this.
Perl Troubleshooting
This is a collection of fixes for various issues I’ve had with Perl. Feel free to add any of your Perl tips here. I will move this to a wiki page if it gets too big.
All Perl scripts fail with error message “Compilation failed in require at…*.pm did not return true at …”
Unable to install packages on Debian with error message “Perl may be unconfigured”
Ph*cking phenolics!
Hi guys,
I have extracted sunflower DNA a dozen times now. I get one consistent problem a gelatinous film coprecipitating with my DNA (carbohydrates/polysaccharaides). I also get sharp smelling phenolic compounds. These occur regardless of DTT/2-BME addition.
I found a paper detailing the use of 2-butoxyethanol (2-BE) to remove strawberry polysaccharides and phenolics. The researcher’s idea is to perform a two-step precipitation:
Silica gel: God’s gift to botanists
Another Rieseberg lab member was asking me this week about preserving plant tissues on silica gel for later DNA extraction, and this got me to thinking about the general idea. I thought I would make a post about it, since it’s a useful technique, even in the genomic age. A lot of you already know all about this, so apologies for preaching to the choir.
I put an extensive post on my own Research Blog, but here is the gist of it:
How to Upload Files to Bitbucket (commandline)
BitBucket is an external source code repository server that hosts our shared code. Our repositories use Git as the version control program to keep track of file changes. The idea is you make changes to code on your local machine then share your code with everyone else by uploading to BitBucket.
The instructions below guide you step-by-step in uploading files to BitBucket using the commandline. Git is one of the most popular version control programs but it is not easy for beginners. If you want to do something that deviates from these steps, consult a git reference. Once you understand the basics of the git workflow, you can use a GUI program which can combine multiple steps in a single click.
“Lab” bike
Summer is almost upon us, and many of you might be considering taking field trips to help out down at Totem or the Farm. So I’d thought I’d let you know about my crappy field bike, available for use to get yourself around campus!
Chemical safety: a quick read
Chemical safety: a quick read
Hi everyone,
Here’s a sobering blog post I saw at BiteSizeBio. I use most of these chemicals regularly. Scarier still I had no idea chloroform becomes phosgene once metabolized. The list contains popular chemicals in the Rieseberg lab.
original link: http://bitesizebio.com/articles/ten-bad-chemicals-in-the-lab-and-what-they-do-to-you/
Ten Bad Chemicals In The Lab and What They Do To You!
SPRI beads (almost) for free!
A few weeks ago Kristin posted a nice blog entry on home made SPRI beads, which effectively replace the ridiculously expensive commercial AMPure beads. (Editor’s note: see this post at RLR for more about the AMPure beads). Dan already ordered the ingredients to make them in our lab as well, and when they all arrived I volunteered to prepare and test them. Following are some results. If you don’t feel like reading through the whole post, the bottom line is that they work very well, and that you are very welcome to use them (they are in an aluminium-wrapped Falcon tube in the fridge, with a white “SPRI beads” label).
Normalizing DNA with the Robot
Im not looking to volunteer myself as the robot expert, this is just one of the things you need if you are going to use it. These are the files needed to normalize a plate of DNA. One instructs it to take the volumes of DNA from one plate and put them into another and the other is to add the water. I can also offer a poor picture of the machine itself.
Freeze and squeeze DNA extraction from gel
Hi all,
Here’s a really easy DNA gel extraction:
1. Go about cutting out your bands of interest in the usual way.
2. Next cut the gel band(s) into tiny cubes.
3. Place tiny cubes in an eppie.
4. Put eppie in -20oC for 15 to 30 min. The idea is to make the band freeze into a mini ice cube.
5. Knock out all of the gel band/cubes onto a 10 (long) x 5 (wide) cm piece of parafilm.
6. Fold the parafilm in half along the length while keeping your gel cubes along the fold in the parafilm. You should have a piece of parafilm folded lengthwise with gel cubes sandwiched between two layers of plastic.
7. Crush/squeeze the liquid out of the tiny cubes. Liquid containing electrophoresis buffer and DNA will separate from the agarose.
8. Collect DNA/electrophoresis buffer mixture with pipet and clean it by ethanol precipitation.
EDIT: Borate in the electrophoresis buffer is a ligase inhibitor. Make sure to thoroughly wash your DNA with 70% EtOH, at least 2 times.
Herbarium Vouchers
In a recent lab meeting, the issue of herbarium vouchering came up, and a spirited discussion ensued. Inspired by what I heard there, and by some discussions with the manager of the UBC Herbarium, I decided to create a post on the how and why of herbarium vouchering. I put the complete version of this on my own research blog, accessible here. Here, I mostly wanted to summarize the idea, and try to convince everyone in the lab that vouchering is really important.
More CTAB!
Since I had as well troubles to get DNA extractions working with the protocols going around in the lab, I tried out a few published protocols (+ the regular CTAB extraction I used for Arabidopsis), picked the one that performed best in my hands and played around with it a bit.
Here’s the result: Sunflower CTAB DNA extraction protocol v1.2
Momma’s CTAB
Like a few other people in the lab, I’ve been struggling with DNA extractions of late. I’ve tried several methods, including the “columnless” method that a few people are using. But I was having pretty spotty luck. Then, during a bout of methodological soul-searching, I also tried a CTAB protocol that used to be my go-to method. It failed me a few weeks ago, but in a second trial last week, it delivered the goods: loads of very pure, high molecular weight DNA. It might not work for all plants, and there do appear to be situations in which it is not the best option, but if anyone wants to get back to down-home, from-scratch extraction, the way momma used to do it, then give this one a try:
Please note: the results below are for Streptanthus (the plant I’m studying; Brassicaceae); I’m trying it for some Asteraceae right now, and will update the post once I have results.
Momma’s DNA Isolation via CTAB
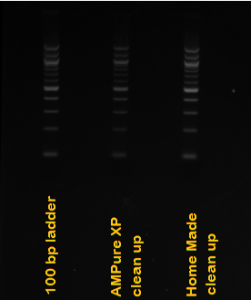
Home Made AMPure XP Beads
AMPure XP beads are used to quickly clean PCR reactions and are also used in WGS library prep. You can also use them to do DNA size selections! They work well, but are very expensive. I found step-by-step instructions on how to make a home-made version online, purchased the reagents and tried the protocol. I tested the beads by attempting to clean up DNA ladder and I found that the home-made beads work identically to the AMPure XP beads.
Attached is the protocol for making the beads. I followed the protocol exactly. One note, I weighed the PEG-8000 in the fume hood because the fine powder is an irritant if inhaled. I also have attached a picture of my gel comparing clean-up of 100 bp DNA ladder with commercial AMPure XP beads and the home-made substitute.
More on Hydroponics
This post is intended to give a few more details on the hydroponics rigs that I constructed, and am currently testing with Greg O. I built these rigs in collaboration with John Gourlay (jgourlay@mail.ubc.ca), who is a technician in the Botany Department workshop (room 1363; directly underneath the room that houses the growth chambers). John was able to create these rigs in less than a day after I described them to him, so if you’re thinking of starting a hydroponics project, you should consider having him do the work for you. He charges a small amount, and does the work very quickly. He’s also a very nice guy.
The rigs are based on a design that is pretty common in the world of hydroponics. The version described below is similar to one developed at Duke University by Jessica Selby and Kevin Wright (John Willis Lab). The idea is to suspend the roots of the plant in a nutrient solution, and provide oxygen to the roots via bubblers. As Greg says, the method requires development. However, it appears to work well for sunflowers (not so much for my plants). Anyway, here is the basic process of constructing the rigs.
Hydroponics test run
Dylan and I have been working on getting hydroponic testing up and running. The basic set up is a large tub filled with dilute nutrient solution, with aeration provided by an air pump. Floating on top of the water is a large foam sheet with holes. In each hole we put a 2 ml tube packed with rock wool and perlite along with a seedling. Extending below the hole is a large bubble tea straw to make sure the roots don’t tangle together while growing.
Methods Write-up for Identifying Contamination in Sequencing Reads
Chris and I devised and implemented this method to identify contaminant DNA in our HA412 454 and Illumina WGS reads.
N.B. I will update this posts with links to the scripts once I get them up on the lab’s bitbucket account, plus extra details if I think they are necessary on review.
distilled water – pH
Picard MarkDuplicates Troubleshooting
Picard is a java-based bioinformatics tools suite that handles SAM/BAM files. Chris introduced me to it, and it’s pretty handy. However, it is very particular about the SAM format, which can leave you searching the FAQs and forums for quite a while.
The MarkDuplicates tool looks for duplicate reads in your library and either flags them in your SAM/BAM file or deletes them, depending on your settings. It can also output a metrics on duplication.
If you are doing any analysis which takes coverage into account, you will want to remove PCR duplicates from your libraries so that they do not artificially inflate coverage numbers. The folks from Celera are adamant about this step, since it can help reduce complexity of genome assembly.
Reads are considered duplicates if they have the same orientation and their 5′ ends map to the same reference coordinate. Picard does not care if reads are part of a pair and either pair end maps to separate sequence entries (e.g separate scaffolds or contigs or chromosomes) in your reference fastas file. This makes it better than samtools rmdup, which is unable to handle read pairs that hit separate reference sequence entries.
Reads marked as duplicate will have the 0x400 bit set in SAM flag (column 2). To check if your read has that bit set, use the Picard Flag Decoder website. When duplicate reads are deleted, the highest quality read is kept intact in the SAM/BAM.