Filtering the variants is a critical step in the pipeline, as most of the SNP callers are very inclusive by default. Below are the criteria I have used to filter SNPs:
SNP calling V – SNP and genotype calling
The next step in the pipeline after the alignments are conducted and adjusted is to call the SNPs and genotypes. There are many programs that will do this such as the ML script that many people in the Rieseberg lab use, freeBayes, mpileup and bcftools, as well as the GATK UnifiedGenotyper. To bench mark the alignment steps in these SNP calling blog posts I used mpileup and bcftools. For example:
SNP calling IV – Base quality score recalibration
The best practices protocol from Broad highly recommends base quality recalibration (http://gatkforums.broadinstitute.org/discussion/44/base-quality-score-recalibration-bqsr). Apparently, the base quality scores off the machine are not very accurate and can provide false confidence in the base calls and consequently influence SNP calling. This tool will adjust average quality scores and also adjust the scores depending on the machine cycle and sequence context (i.e. start versus end of read, type of dinucleotide repeat). After recalibration, the quality score is closer to its actual probability of mismatching the reference genome.
To run this you need a database of known SNPs so the tool can assess the error rate at other sites based on the actual sequence data. Unlike humans, which this tool was designed for, most species do not have comprehensive SNP databases. However, such a table can be created by identifying SNPs in a preliminary run without the base quality score recalibration. I included any SNP that had a Qual score of 20 in my SNP vcf file.
You can see below that it had a large effect on the number of SNPs that I called. With the BAQ filter off 263,360 SNPS overlapped between the methods. 14,591 were unique to the recalibrated alignments and 131,354 were unique to the alignments without base quality recalibration.
SNP calling III – The indel problem
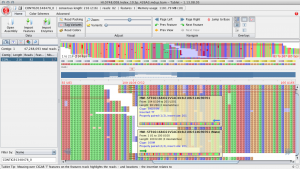
SNPs can often be falsely called around indels. In particular, if indels are near the start or end of a read the read is often incorrectly aligned. Here is an example of how things can go wrong from a spruce alignment. Below, at site 109 there is a TT insertion in the reads relative to the reference and there is no true SNP at this site. However, this individual was called as a het (C/T) because of the reads that end in the middle of the insertion. For these reads the penalty for a single mismatch to the reference is less than introducing a gap.
SNP calling II – Creating a reference for GATK and Picard
Using GATK and Picard was difficult because we had to keep modifying our reference (and therefore redo the alignments each time) to use these programs efficiently. Below I list the things we had to do to the reference to get them to work:
SNP calling I – alignment programs and PCR duplicates
I have tested various SNP calling methods using exome re-sequencing data from 12 interior spruce samples. I tried Bowtie2, BWA (mem), Picard (mark duplicates) and GATK for indel realignment and base quality recalibration. For SNP calling I used mpileup with and without BAQ as well as the Unified Genotyper from GATK. For an interesting and informative workshop outlining the Broad best practices SNP calling pipeline check out these youtube videos (http://www.youtube.com/watch?v=1m0ZiEvzDKI&list=PLlMMtlgw6qNgNKNv5V9qmjAxbkHAZS1Mf). My results are in a series of blog posts and I hope you find them useful. Please let me know if you have any suggestions for SNP calling. We only want to do the alignments and SNP calling once for the entire set of samples, because it is going to take a long time!
Estimating Insert Sizes
We recently had some trouble estimating insert sizes with our Mate Pair (aka Jumping, larger insert sizes) Libraries. All the libraries sequenced by Biodiversity and the Genome Sciences Centre (GSC) were shockingly bad, but the libraries sequenced by INRA were very good. For example, according to the pipeline, the GSC 10kbp insert size library had an average 236bp insert size, but the INRA 20kb library an average insert size of 20630bp.
See the histogram for the 10kbp library:
GBS snp calling
One of the main ways we call SNPs in GBS data is using a maximum likelihood script derived from the original Hohenlohe stickleback RAD paper. It uses a likelihood function based on the number of reads for each base at a site to determine if its homozygous or heterozygous.
It’s been known for a while that it seemed to be biased against high coverage heterozygous sites. I took the script apart and realized that there was actually a serious problem with it. The likelihood function used factorials, and when there were over 175 reads at a site, perl broke down and started describing things as infinite. Then the script would call it as a homozygote by default.
Letting perl work with the huge numbers necessary makes it incredibly slow, so I put in a logic gate for high coverage sites. If there is over 150 reads at a site, and the second most numerous base makes up atleast 10% of the reads (i.e. at minimum 15), then it is heterozygous. Otherwise it is homozygous.
The rest of the script is the same. One could debate how best to call the high coverage scripts, but I suggest you use this script over the old one.
Extracting point data from vector and raster maps (for free)
I recently wrote a post on my research blog all about using free GIS software to extract point data from vector and raster maps. I won’t repeat it here, but this is the upshot:
One of the most powerful uses of GIS technology is to sample and analyze data from GIS data layers. For instance, if you have a set of GPS coordinates for a species of plant from Utah, and you want to find out what range of climate types the plant grows in, you can use digital raster layers for climate (high and low temperature, rainfall, etc.) to extract estimates for the conditions that your plants experience at the locations where you sampled it. Then, you can use that data for many purposes: reconstruct it on a phylogenetic tree, test for divergence in climate regime among populations, test for associations among variable (for instance among soil and climate). Really, anything.
Let me know if you have any questions! I’m happy to help people get this going for specific data sets.
Filtering SNP tables
Previously I posted a pipeline for processing GBS data. At the end of the pipeline, there was a step where the loci were filtered for coverage and heterozygosity. How strict that filtering was could be changed, but it wasn’t obvious in the script and it was slow to rerun. I wrote a new script that takes a raw SNP table (with all sites, including ones with only one individual called) and calculates the stats for each loci. It is then simple and fast to use awk to filter this table for your individual needs. The only filtering the base script does is not print out sites that have four different bases called at the site.
Heterozygosity, read counts and GBS: PART 2
Subtitle: This time… its correlated.
Previously I showed that with the default ML snp calling on GBS data, heterozygosity was higher with high and low amounts of data. I then took my data, fed it through a snp-recaller which looks for sites that were called as homozygous but had at least 5 reads that matched another possible base at that position (i.e. a base that had been called there in another sample). I pulled all that data together, and put it into a single table with all samples where I filtered by:
Lab Safety training
UBC offers two levels of safety courses.
For those who work under supervision (i.e. undergrads), there is a basic online course found here http://riskmanagement.ubc.ca/courses/intro-lab-safety
This is a requirement for all undergrads who will be working in the lab. Once your student has taken the course, it is expected that you (their supervisor) will follow up with reviewing lab safety training specific to our lab. Please remember that these students need supervision at all times when working in the lab.
For lab personnel who do not require supervision there is a more advanced course, http://riskmanagement.ubc.ca/courses/laboratory-chemical-safety, it includes an online and practical component. This course is mandatory, unless you’ve had similar training at a previous institution. Megan will provide in lab follow-up for new personnel to cover safety features of the lab.
UBC also offers other specific training, although the courses aren’t likely relevant to the type of lab work we do; such as Radionucleotides, and biohazards, please see http://riskmanagement.ubc.ca/courses.
C/C++ Dependency Troubleshooting
Unix C/C++ programs are very finicky about the compiler and library versions. Compiling is the process of translating human readable code into a binary executable or library that contains machine-friendly instructions. Linking is the process of telling multiple executables or libraries how to talk to each other.
gcc is a GNU C compiler that is typically used on unix systems. g++ is a GNU C++ compiler on unix systems. libstdc++ is the GNU standard C++ library. glibc is the GNU standard C library. When you install GCC on your unix machine, you are installing a package of the aforementioned items. The gcc unix command is smart enough to call either the gcc compiler or g++ compiler depending if you pass it C or C++ code.
If you attempt to run your program with an older standard library than it was originally linked it with, your program will crash and complain. Here are some tips to get around it.
C / C++ Troubleshooting
IN PROGRESS
Most of the high-performance bioinformatic programs are written in C or C++. Unfortunately, C and C++ code is some of the hardest code to debug. If you have only programmed casually in perl/python, you will not have a good time. Here are some tips to help you out, but you will most likely need someone with C / C++ programming experience and knowledge of the code to get you through it.
SmartmonTools & GSmartControl
Smartmontools is a command-line Hard Drive Diagnostic Tool that gives you clues on how long your disk has to live. You can run it manually, or you can configure it to periodically test your drives in the background and notify you about test failures via email.
GSmartControl is a GUI for Smartmontools and much easier to use.
Check out this Ubuntu SmartmonTools Tutorial on how to install and set them up.
Here are some tips that are not easily gleaned from the previous websites:
Continue reading
Nice Sunflower Pics
Saw this link with some nice (and some marginally nice) photos of Helianthus spp:
http://earthsky.org/earth/sunflowers-see-a-dozen-great-photos
AWK unlinked SNP subsampling
For some analyses, like STRUCTURE, you often want unlinked SNPs. For my GBS data I ended up with from 1 to 10 loci on each contig which I wanted to subsample down to just one random loci per contig. It took me a while to figure out how to do this, so here is the script for everyone to use:
cat YOUR_SNP_TABLE | perl -MList::Util=shuffle -e ‘print shuffle(<STDIN>);’ | awk ‘! ( $1 in a ) {a[$1] ; print}’ | sort > SUBSAMPLED_SNP_TABLE
It takes you snp table, shuffles the rows using perl, filters out one unique row per contig using awk, then sorts it back into order. For my data, the first column is CHROM for the first row and then scaffold###### for the subsequent rows so the sort will place the CHROM row back on top. It might not for yours if you have different labels.
GBS, coverage and heterozygosity
I’m running some tests on my GBS data to look for population expansion. I know from looking at GBS data from an F1 genetic mapping population that for GBS data heterozygotes can be under called due to variation in amplification and digestions. Also, for my data observed heterozygosity is almost always under expected. Heterozygotes can also be overcalled when duplicated loci are aligned together. The tests I’m going to use explicitly use observed heterozygosity so this is worrying.
Text Editor Indenting & Highlighting
Do you hate reading code that is poorly indented or lacks syntax highlighting? Here are some common text editor fixes:
Continue reading
Batch Sequence Read Archive Submissions
Getting all of our data uploaded to the SRA is important. It is good to share our data publicly whenever possible, and just as importantly, it provides us with a free off-site backup of our raw data.
We keep a spreadsheet of the lab’s sequencing data here: http://bit.ly/17Z4X1P
*IMPORTANT*: The spreadsheet is curated by a few members of the lab and is not complete. Your data may not be listed here, if it is not, please do your best to add it. You can email me and Sebastien with any questions you might have.
I’ve added a column that indicates the submission status of each sample in the Sunflower Relatives and Wild Sunflowers tabs, so you can tell if your data has already been submitted.