We recently had some trouble estimating insert sizes with our Mate Pair (aka Jumping, larger insert sizes) Libraries. All the libraries sequenced by Biodiversity and the Genome Sciences Centre (GSC) were shockingly bad, but the libraries sequenced by INRA were very good. For example, according to the pipeline, the GSC 10kbp insert size library had an average 236bp insert size, but the INRA 20kb library an average insert size of 20630bp.
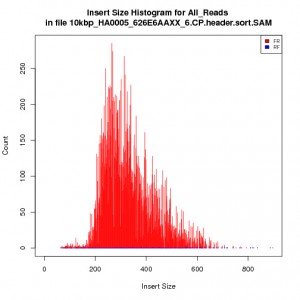
See the histogram for the 10kbp library:
In our pipeline, we aligned whole genome shotgun Helianthus annuus reads against a Helianthus annuus chloroplast reference found on NCBI and fed the SAM alignments into Picard CollectInsertSizeMetrics.
The discrepency between the expected and actual insert sizes for non-INRA libraries were just too horrible, indicating that there was an issue with the pipeline. What was weird was that all libraries were run through the same pipeline with the same reference. Also, the same pipeline did fine estimating insert sizes for Paired End (aka Fragment, smaller insert size) libraries from GSC & Biodiversity.
Chris found that Picard CollectInsertSizeMetrics automatically trims the histogram width to 10 x standard deviation to eliminate chimeras and other bad sequencing from affecting the insert size estimation. Once we explicity set the histogram width to a number much larger than the expected insert size (40 000), Picard reported much more reasonable insert sizes for the GSC and Biodiversity libraries. The problem was that the GSC and Biodversity long-insert libraries were poor quality and had a lot of mini fragments around 200-300bp long. Although the expected insert size was within 10 X standard deviations of the actual average insert size, Picard was removing the larger insert size reads from the statistics report. Most likely, Picard does not calculate the average insert size using all reads, otherwise the larger insert size reads would have been kept.
See here for the more accurate insert size statistics for the same 10kbp library:
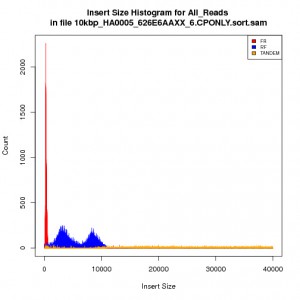
10kbp insert size library Picard CollectInsertSizeMetrics histogram with histogram width explicity set
These were the Picard commands that we used:
BAD:
java -Xmx2g -Djava.io.tmpdir=/data/insertsize/tmp -jar CollectInsertSizeMetrics.jar HISTOGRAM_FILE=10kbp.hist.pdf INPUT=10kbp.sort.sam OUTPUT=10kbp.picard.insert_size_metrics VALIDATION_STRINGENCY=LENIENT REFERENCE_SEQUENCE=cp.descriptive_headers.fasta MINIMUM_PCT=0 &> 10kbp.picard.log
GOOD: Note that the histogram width is explicity set to a number much larger than the expected insert size of 10kbp
java -Xmx2g -Djava.io.tmpdir=/data/insertsize/tmp -jar CollectInsertSizeMetrics.jar HISTOGRAM_FILE=10kbp.hist.pdf INPUT=10kbp.sort.sam OUTPUT=10kbp.picard.insert_size_metrics VALIDATION_STRINGENCY=LENIENT REFERENCE_SEQUENCE=cp.descriptive_headers.fasta MINIMUM_PCT=0 HISTOGRAM_WIDTH=40000 &> 10kbp.picard.log