I have been trying for months to find a high-throughput solution to DNA extraction from lyophilized (freeze-dried) leaf discs of Helianthus argophyllus, a species known for its intransigent polyphenolics and low DNA yields. And I appear to have found one, thanks to Horne et al. 2004 (here) and Dow Chemical!
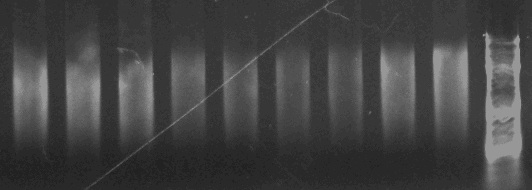
The protocol (my version linked here: modified Qiagen protocol) is a modified 96-well Qiagen plant DNA extraction kit. It takes about an hour longer, and involves a bit more preparation, but at least for me the protocol is extremely effective. On a gel, DNA I obtained with the non-modified 96-well Qiagen kit looked like this:
With an average nanodrop estimated concentration of ~13 nanograms/microliter and 260/230 of ~1.5 to 1.7. Low and dirty.
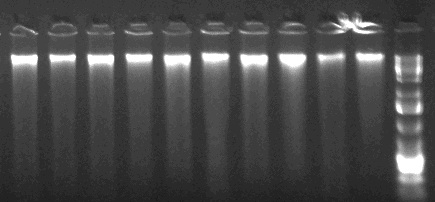
With the modified protocol my DNA looks like this:
It is unsheared, despite having been put through a column. It appears that mechanical shearing occurs via pipetting, and so I strongly recommend using cut tip pipette tips during the supernatant transfer step. DNA from this protocol has an average estimated concentration of ~22 nanograms/microliter and 260/230 of ~2.05 to 2.15. SO MUCH BETTER!