Congratulations to lead author Quinn McCallum on today’s publication of his paper on genomic differentiation of Zonotrichia sparrows in the Journal of Evolutionary Biology. Quinn conducted this work while an Honours undergraduate student at UBC. Quinn and I are also grateful for the contributions of coauthors Kenny Askelson, Finola Fogarty, Libby Natola, Ellen Nikelski, and Andrew Huang. We also thank WildResearch and the Iona Island Bird Observatory for their contributions to the study.

The abstract:
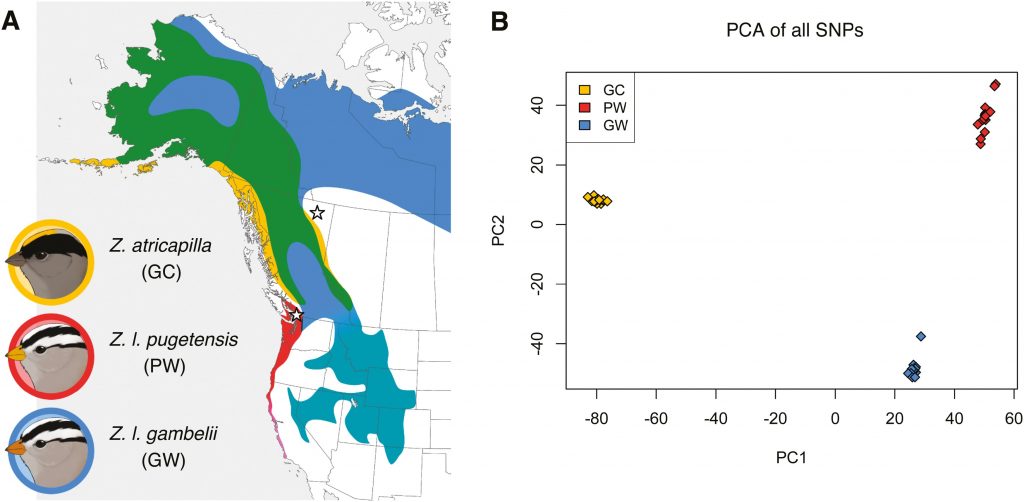
When a single species evolves into multiple descendent species, some parts of the genome can play a key role in the evolution of reproductive isolation while other parts flow between the evolving species via interbreeding. Genomic evolution during the speciation process is particularly interesting when major components of the genome—for instance, sex chromosomes vs. autosomes vs. mitochondrial DNA—show widely differing patterns of relationships between three diverging populations. The golden-crowned sparrow (Zonotrichia atricapilla) and the white-crowned sparrow (Zonotrichia leucophrys) are phenotypically differentiated sister species that are largely reproductively isolated despite possessing similar mitochondrial genomes, likely due to recent introgression. We assessed variation in more than 45,000 single nucleotide polymorphisms to determine the structure of nuclear genomic differentiation between these species and between two hybridizing subspecies of Z. leucophrys. The two Z. leucophrys subspecies show moderate levels of relative differentiation and patterns consistent with a history of recurrent selection in both ancestral and daughter populations, with much of the sex chromosome Z and a large region on the autosome 1A showing increased differentiation compared to the rest of the genome. The two species Z. leucophrys and Z. atricapilla show high relative differentiation and strong heterogeneity in the level of differentiation among various chromosomal regions, with a large portion of the sex chromosome (Z) showing highly divergent haplotypes between these species. Studies of speciation often emphasize mitochondrial DNA differentiation, but speciation between Z. atricapilla and Z. leucophrys appears primarily associated with Z chromosome divergence and more moderately associated with autosomal differentiation, whereas mitochondria are highly similar due apparently to recent introgression. These results add to the growing body of evidence for highly heterogeneous patterns of genomic differentiation during speciation, with some genomic regions showing a lack of gene flow between populations many hundreds of thousands of years before other genomic regions.
The citation:
McCallum, Q., K. Askelson, F.F. Fogarty, L. Natola, E. Nikelski, A. Huang, and D. Irwin. 2024. Pronounced differentiation on the Z chromosome and parts of the autosomes in crowned sparrows contrasts with mitochondrial paraphyly: implications for speciation. Journal of Evolutionary Biology, voae004, https://doi.org/10.1093/jeb/voae004
Quinn is now a PhD student in the Mason Lab at Louisiana State University. You can learn more about him and his work here.