 Genomic history of invasion
Genomic history of invasion
Biological invasions of non-native species present compelling motivation to understand how human-induced changes in the environment and species distributions influence ecological and evolutionary processes. Their documented geographic spread across time makes them ideal for study using historic collections, allowing better insight to evolutionary change over short time scales. Applying advanced genomic approaches to historic samples is key to understanding the processes that allow plants to rapidly establish and adapt to new environments. Theory predicts that dramatic ecological and evolutionary changes affect invasive species soon upon arrival in a new habitat. Yet current research relies on sampling contemporary populations, and therefore reveals little about the initial stages of invasion. Here, we study of the history of an invasive weed by exploiting an untapped historical resource to observe “snapshots” of the initial stages of invasion and the genetic changes that occur as a plant species spreads. It involves sampling genetic material from dried plant specimens collected throughout the course of an invasion, from herbarium collections across North America. Techniques for ancient DNA originally developed to study long extinct organisms such as mammoths are being used to study evolution over the course of the 100 year invasion of North America by crossflower (Chorispora tenella, Brassicaceae), a widespread and governmentally listed noxious invasive weed.
Collaborators: Ruth Hufbauer, John McKay, Hernán Burbano, Rafal Gutaker

Genetics of embryo development in an early branching Brassicaceae
Crossflower is located in a clade sister to the rest of the Brassicaceae (the Chorisporeae), with several interesting and rare reproductive developmental traits among the Brassicaceae, including segmented fruit and wingless seeds (Smith et al. 2011; Huang et al. 2015). In addition to crossflower, the Brassicaceae offers a wealth of comparable systems with available reference genomes and well-described seed and fruit development, and boasts some of the best work in the area of evolution of development, thereby allowing easy identification of parallels and divergences presented by crossflower (Koenig & Weigel 2014). The extent of previous genomic work in this family suggests the potential to zoom into the gene level of information and take advantage of existing annotation and functional information. For example, within the Brassicaceae, only Chorispora and members of the hugely economically important Brassiceae tribe (which contains canola, cabbage, and radish) demonstrate fruit segmentation, a type of shattering (Huang et al. 2015). In an oilseed crop such as canola this trait is extremely undesirable and a major cause of yield loss, and so extensive effort has focused on understanding the genes and regulatory pathways involved, typically using Arabidopsis (which has non-segmented fruits) as a model. With the genomic data generated by this project, and the collaboration of developmental geneticist with expertise in the Brassicaceae, I will be able to compare orthologous genes in Chorispora and perhaps gain a better understanding of embryo development and fruit segmentation. These genes may play an important role in seed dispersal (as shown in Arabidopsis; Liljegren et al. 2000) and thereby contribute to the spread of this invasive crop weed across North America in less than 100 years.
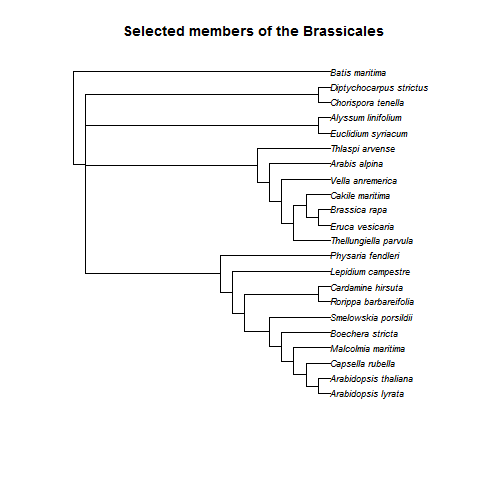
Collaborators: Ruth Hufbauer, John McKay, Martin Bayer
Training for this project partially funded by microMORPH.
