Lecture 2
Gene Structure
MOLECULAR DEFINITION OF A GENE
In the past, the term gene was used to refer only to a DNA sequence that was transcribed into RNA. Today's definition includes all the sequences transcribed into RNA (introns and exons) and the proximal and distal sequences which control the expression of the gene (the promoter, proximal- promoter control sequences and distal enhancers).

Basic Definitions:
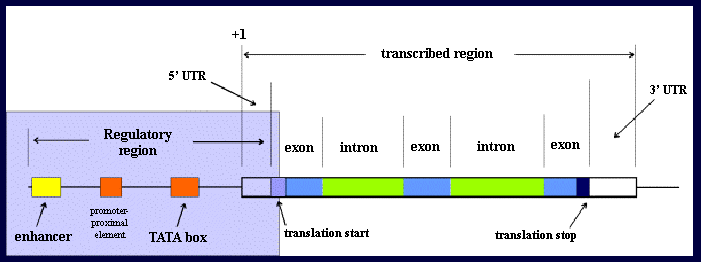
Transcription start: Base 1 of the primary transcript
Transcription stop: Where the RNA polymerase falls off the DNA
Translation start: The AUG (Methionine) codon
Translation stop: The termination codon (TAA, TGA, TAG)
Polyadenylation site: 3' end of the mRNA
5' untranslated region: Between base 1 and the AUG, often called the leader
3' untranslated region: Between the termination codon and polyadenylation site
5' flanking region: From -1 continuing upstream in a 5' direction
3' flanking region: Usually from polyadenylation site continuing downstream in 3' direction
Regulatory region: Refers to enhancer and promoter when these have yet to be characterized.
Blackwood and Kadonaga (1998). Science 281, 60-63
More definitions:
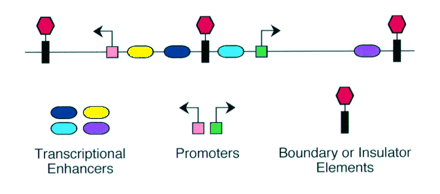
Enhancer - A type of regulatory sequence in eukaryotic DNA (rarely in prokaryotes) that may be located at a great distance upstream or downstream from the promoter which it influences. Binding of specific proteins to an enhancer stimulates or decreases (silencer) the rate of transcription of the gene. Features thought to be unique to enhancers are that they function in either orientation and can function 5', 3' or within the gene. Enhancers work by binding transcription factors, of which we will see much more in future lectures. Transcription factors either have activating or repressive domains, or recruit activator or repressor proteins, which in turn influences gene expression.
Blackwood and Kadonaga (1998). Science 281, 60-63
Promoter - DNA sequence which determines the site of transcription initiation for a RNA polymerase and is required for basal transcription. Most promoters contain several sequences or motifs that contribute to promoter function. For example, the CCAAT box and the TATA box.
CCAAT box (-80 to -70) - Binds a family of related transcription factors highly conserved in evolution. Usually considered part of the promoter because full level of basal transcription is not achieved in its absence.
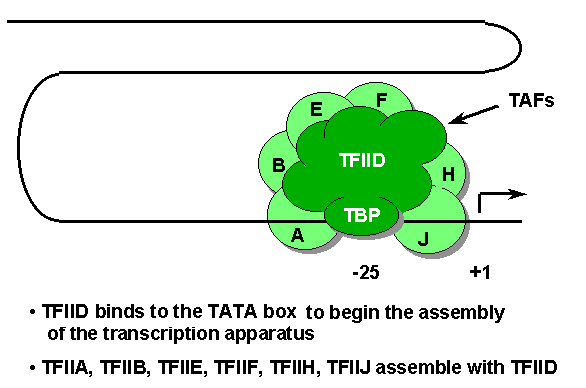
TATA box (-30 to -20) - The TATA box is a component of many promoters utilized by RNA polymerase II. The first step in the assembly of a transcription initiation complex, a subunit of TFIID called TBP (TATA-binding protein) binds the TATA box. Once TFIID is bound, the other general transcription factors along with RNA polymerase II, are added in turn.
Initiator element (Inr) - The Inr was originally identified as a sequence that encompasses the transcription start site that is sufficient to direct accurate initiation in the absence of a TATA element. Inr elements are, however, present in both TATA-containing and TATA-deficient (TATA-less) promoters.
Downstream promoter element (DPE) - The downstream promoter element (DPE) functions cooperatively with the initiator element for the binding of TFIID in the transcription of core promoters in the absence of a TATA box.
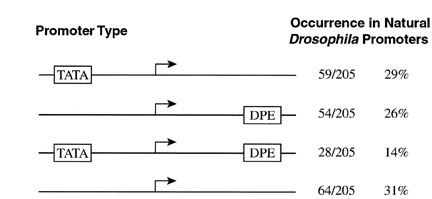
Kutach and Kadonaga (2000). MCB 20, 4754-64.
Promoter-proximal elements - Any regulatory sequence in eukaryotic DNA that is located close to (within 200 base pairs) a promoter and binds a specific protein thereby modulating transcription of the associated protein coding gene. Many genes are controlled by multiple promoter-proximal elements.
Transcription control region - collective term for all the cis-acting DNA regulatory sequences that regulate transcription of a particular gene.Boundary or Insulator Elements - DNA segment from about 0.5 to 3kb that are thought to function as transcriptionally neutral DNA elements that block, or insulate, the spreading of the influence of nearby enhancers or heterochromatin-like repressive effects.
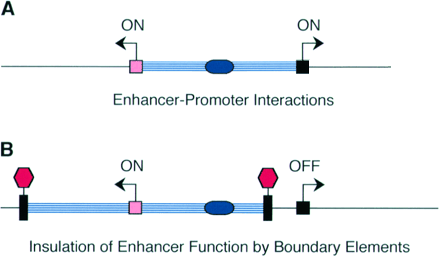
Modified from Blackwood and Kadonaga (1998). Science 281, 60-63
Transcription factors - general term for any protein, other than RNA polymerase, required to initiate or regulate transcription in eukaryotic cells. General factors required for transcription of all genes, participate in the formation of the transcription initiation complex near the start site. Specific factors stimulate (or in some cases repress) transcription of particular genes by binding to their regulatory sequences (eg. enhancers and promoter-proximal elements).
Upstream activating sequence (UAS) - any protein-binding regulatory sequence in the DNA of yeast and other simple eukaryotes that is necessary for maximal gene expression; UAS is functionally equivalent to an enhancer in higher eukaryotes.