Protein Targeting - Nucleus, Mitochondria
and Chloroplasts.
1. Targeting signals
All proteins have a common beginning.
Synthesis of all proteins, regardless of their final destination is initiated
is initiated on free ribosomes in the cytoplasmic compartment of the cell.
Any differences in the fate of proteins is a consequence of targeting signals
contained within the amino acid sequence of the protein itself.
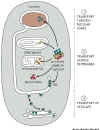
Figure 14-5
|
Key concept:
Each protein is destined by its amino sequence to go to certain places and
to do certain things. Different proteins contain different addressing
information that enables them to be routed to different specific destinations.
See Fig 14-5.
Changes in amino acid sequence (mutations)
often result in improper transport and processing of proteins so that
the abnormal polypeptides do not appear in the normal position for the
protein.
|
There are three major types of transport systems
- Nuclear pores
- Translocation across membranes.
- Vesicle transport
The key problem is how to get largely hydrophilic
proteins across the hydrophobic barrier presented by the membrane core. -- It
is done in the same way that transporters for smaller hydrophilic molecules
work, by creating a controlled hydrophilic passage through the membrane
for specific molecules.
Signal information is the key. It corresponds
to the address on a letter and is contained in the amino acid sequence of the
protein. It allows the protein to be regognized and processed by other proteins.
Table
14-3 Some Typical Signal Sequences
|
| Function of Signal |
Example of
Signal Sequence |
|
| Import
into ER |
+H3N-Met-Met-Ser-Phe-Val-Ser-Leu-Leu-Leu-Val-Gly-Ile-Leu-Phe-Trp-Ala-
Thr-Glu-Ala-Glu-Gln-Leu-Thr-Lys-Cys-Glu-Val-Phe-Gln- |
| Retention
in lumen of ER |
-Lys-Asp-Glu-Leu-COO- |
| Import
into mitochondria |
+H3N-Met-Leu-Ser-Leu-Arg-Gln-Ser-Ile-Arg-Phe-Phe-Lys-Pro-Ala-Thr-Arg-
Thr-Leu-Cys-Ser-Ser-Arg-Tyr-Leu-Leu- |
| Import
into nucleus |
-Pro-Pro-Lys-Lys-Lys-Arg-Lys-Val- |
| Import
into peroxisomes |
-Ser-Lys-Leu- |
Positively charged amino acids are shown in red,
and negatively charged amino acids in green.
An extended block of hydrophobic amino acids is shown in blue.
+H3N indicates the amino terminus of a protein;
COO- indicates the carboxyl terminus.
|
In general, the signal sequences are located
at or near the amino-terminal end of the polypeptide. Notice in the table above
that the exception is the ER retention signal, which is located at the C-terminal
end of the protein.
Each of the signal sequences has a characteristic
amino acid motif: for example, in the nuclear localization signal the key motif
is lys-lys-lys-arg-lys.
The signal sequences are critical for proper
targeting of proteins.
Key experiment: Take the signal peptide
sequence from a protein that is normally targeted to the lumen of the ER and
put it onto the N-terminal end of a protein that normally is found in the cytosol:
The modified protein is now located in ER. Conversely, if the signal sequence
is removed from the protein that is normally targeted to the ER lumen, it does
not enter the ER, but stays in the cytosol.
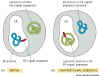 |
Figure 14-6 experimental
test of the role of signal sequences. IMPORTANT |
2. Entering the nucleus.
Good link for nuclear
pores. Lots of material
Entry of large proteins into
the nucleus requires:
- Nuclear Localization
Signal (NLS) lys-lys-lys-arg-lys.
inside the protein, near but not at the N-terminal end of protein.
- NLS region of protein binds
to a cytosolic nuclear import receptor protein (NIR)
- NIR with attached NLS-protein
then binds to the nuclear pore. A GTP driven reaction results in a change
in the configuration of the pore that results in the translocation (movement)
of the NIR-NLS-protein complex into the nucleus
- When there, the NIR proteins
dissociate from the NLS-protein and are returned to the cytosol.
- As the NLS is an internal
localization signal it is not removed by a specific peptidase as are
ER, mitochondrial and chloroplast localization signals that are located at
the N terminal end of the protein.
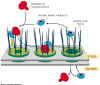 |
Figure 14-9 Import of proteins into the nucleus. Notice the
role of the nuclear import receptor protein (blue). Link
to animation. |
3. Entry to
a mitochondrion, chloroplast or peroxisome
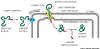 |
Key concept:
Protein containing signal sequence is recognized, bound, unfolded on entry
and refolded on the organellar side of the membrane. See Fig 14-10 (left).
This requires a set of special organellar chaperone proteins. |
The protein recognition signal is quite large
(See table). The entry site to mitochondria or chloroplasts
is a site where the inner and outer membranes touch. This is referred to as
a contact site.
- The protein containing the signal sequence
is synthesized in the cytoplasm.
- Signal sequence binds to a receptor in
the organelle membrane
- Receptor - protein complex diffuses within
membrane to a contact site.
- Protein is unfolded, moved across the membrane,
and refolded. These operations are carried out by the protein transporter
complex and its associated chaperone
proteins. Remember chaperones from earlier discussion
of protein processing? The signal sequence is the first part of the
protein to enter the organelle.
- Once inside, the signal sequence is
cleaved off by a specific peptidase.


