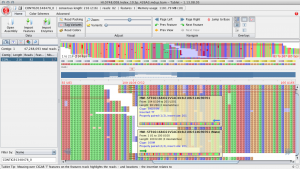
SNPs can often be falsely called around indels. In particular, if indels are near the start or end of a read the read is often incorrectly aligned. Here is an example of how things can go wrong from a spruce alignment. Below, at site 109 there is a TT insertion in the reads relative to the reference and there is no true SNP at this site. However, this individual was called as a het (C/T) because of the reads that end in the middle of the insertion. For these reads the penalty for a single mismatch to the reference is less than introducing a gap.
Tag Archives: Picard
SNP calling II – Creating a reference for GATK and Picard
Using GATK and Picard was difficult because we had to keep modifying our reference (and therefore redo the alignments each time) to use these programs efficiently. Below I list the things we had to do to the reference to get them to work:
SNP calling I – alignment programs and PCR duplicates
I have tested various SNP calling methods using exome re-sequencing data from 12 interior spruce samples. I tried Bowtie2, BWA (mem), Picard (mark duplicates) and GATK for indel realignment and base quality recalibration. For SNP calling I used mpileup with and without BAQ as well as the Unified Genotyper from GATK. For an interesting and informative workshop outlining the Broad best practices SNP calling pipeline check out these youtube videos (http://www.youtube.com/watch?v=1m0ZiEvzDKI&list=PLlMMtlgw6qNgNKNv5V9qmjAxbkHAZS1Mf). My results are in a series of blog posts and I hope you find them useful. Please let me know if you have any suggestions for SNP calling. We only want to do the alignments and SNP calling once for the entire set of samples, because it is going to take a long time!
Estimating Insert Sizes
We recently had some trouble estimating insert sizes with our Mate Pair (aka Jumping, larger insert sizes) Libraries. All the libraries sequenced by Biodiversity and the Genome Sciences Centre (GSC) were shockingly bad, but the libraries sequenced by INRA were very good. For example, according to the pipeline, the GSC 10kbp insert size library had an average 236bp insert size, but the INRA 20kb library an average insert size of 20630bp.
See the histogram for the 10kbp library: