


DNA - Molecule that encodes the genetic information of an organism. Consists of a sequence of nucleotides: Adenine, Cytosine, Guanine, Thymine).

Chromosome - Composed of DNA (in a linear or a circular arrangement) and associated proteins and RNA.


Gene - Segment of the DNA, often one that codes for a specific protein.

Allele - A variant of a gene (eg A, B, or 0 in the ABO blood system).





...but it also destroys beneficial gene combinations.
...but it can also slow it down.
...but it can also reduce it.
 Under what conditions is recombination favorable?
Under what conditions is recombination favorable?
 When will evolution lead to increased recombination rates?
When will evolution lead to increased recombination rates?


For simplicity, I will consider a population of haploids where adults carry only one copy of each chromosome (examples in algae, fungi, protists, and bacteria).
Life Cycle:

Gamete union: Assumed to occur at random.
Selection: Assumed to depend on genotype at two genes (A and B).

Model 1: Beneficial alleles. No fitness interactions between loci.
Model 2: Deleterious alleles. Fitness interactions between loci (measured by  ).
).


Mutation rates to beneficial alleles are generally smaller than to deleterious alleles.
Model 1: Beneficial mutation arises in a single copy and we examine its fate (stochastic analysis).
Model 2: Mutations happen continuously at a rate 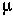 (deterministic analysis).
(deterministic analysis).
Recombination: Occurs between the two genes (A and B) at a rate r.
This rate may be fixed or it may depend on the alleles carried a third gene (M) that modifies recombination rates:

 Such modifier loci are used to investigate how a trait will evolve.
Such modifier loci are used to investigate how a trait will evolve.
The modifier gene recombines with the nearest selected gene (A) at a rate R.


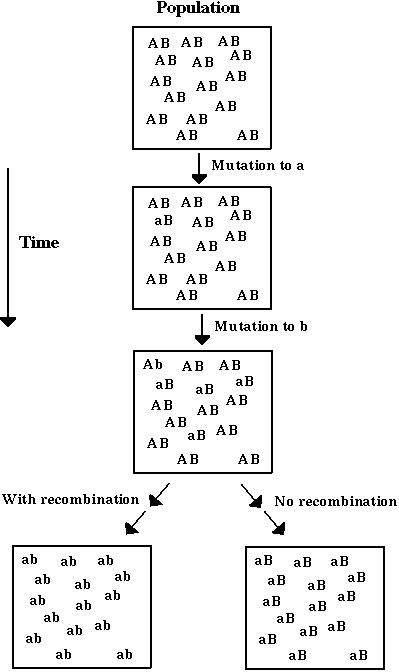
Dynamically, recombination acts to randomize the alleles carried at different genes on a chromosome.
That is, recombination acts to reduce genetic associations between genes. These associations are measured by the linkage disequilibrium (D):


In Model 1, genetic associations are created as beneficial mutations just happen to occur on a particular chromosome.
In Model 2, genetic associations are created because of the fitness interactions between the two genes.


For, unless advantageous mutations occur so seldom that each has had time to become predominant before the next appears, they can only come to be simultaneously in the same gamete by means of recombination.


Calculating the Probability of Fixation: We assume that an individual carrying a new beneficial mutation has a mean number of offspring (1+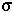 ) that is greater than the replacement rate (1) and that the offspring distribution is Poisson.
) that is greater than the replacement rate (1) and that the offspring distribution is Poisson.
The fixation probability of a beneficial allele that arises at time t is:
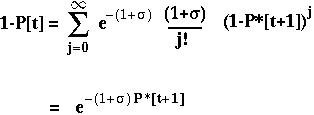
P*[t+1] is the expected fixation probability of a single copy of the allele in an offspring.
With only one gene, P*[t+1] equals P[t] and
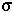 equals s:
equals s:
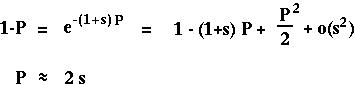
(Classical result from Haldane 1927.)


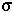 , depends on which allele it appears with at gene B.
, depends on which allele it appears with at gene B.
Furthermore, the fixation probability of an offspring copy, P*[t+1], is no longer equal to the fixation probability of the parental copy, since recombination may place the mutant allele onto a different genetic background.
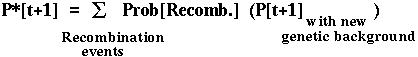
Since there are four genetic backgrounds (MB,Mb,mB,mb), this approach leads to four equations describing how the probability of fixation of an allele changes over time.
These equations can be solved numerically and can be approximated analytically (by assuming that selection is weak and replacing the difference equations with differential equations).


 Barton (1995) showed that recombination always increases the fixation probability of a beneficial mutant in this model.
Barton (1995) showed that recombination always increases the fixation probability of a beneficial mutant in this model.
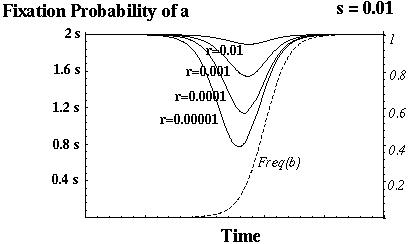
 Otto & Barton (1996) showed that modifier alleles that increase recombination always increase in frequency.
Otto & Barton (1996) showed that modifier alleles that increase recombination always increase in frequency.
 The selection for increased recombination is only strong, however, if the genes are tightly linked or if there are beneficial alleles segregating at many different genes.
The selection for increased recombination is only strong, however, if the genes are tightly linked or if there are beneficial alleles segregating at many different genes.


For theoretical reasons (with some experimental support), it is thought that deleterious mutations may interact such that their combined effects are worse than expected on the basis of their effects singly.
 Negative epistasis (
Negative epistasis ( < 0).
< 0).
If a and b are the deleterious alleles, the fact that there is negative epistasis means that selection acts strongly against the ab combination, which is consequently rarer than expected.
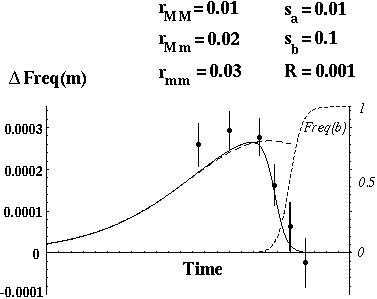
In other words, negative disequilibrium develops when there is negative epistasis (shown by Eshel and Feldman 1970).
With negative disequilibrium in the population, a modifier allele (m) that increases recombination will increase the frequency of the underrepresented genotypes (AB and ab).
This has two consequences:

If epistasis is strongly negative, the modifier allele m will decrease the average fitness of the offspring.


By performing a local linear stability analysis of these recursions it can be shown that

This condition is easily met if epistasis is very weak (relative to selection), but is difficult to meet when epistasis is strong.
(Otto and Feldman 1996)
 Negative epistasis favors the evolution of recombination, but only if the modifier gene is tightly linked (small R) or if fitness interactions are weak.
Negative epistasis favors the evolution of recombination, but only if the modifier gene is tightly linked (small R) or if fitness interactions are weak.


Modeling evolutionary processes allows us
Two hypotheses for the evolution of recombination were confirmed in this work, but found to have greater restrictions than previously thought.
but the strength of selection for increased recombination is only substantial if genes are tightly linked or if there are many beneficial mutations in the genome.
Increased recombination can evolve...
but only if the genes are tightly linked or if the fitness interactions are weak.