Modeling X-chromosome inactivation in human blood cells
Introduction
X-chromosome inactivation (XCI) is a process that occurs at an early stage of development in the human body. At the time of XCI, a certain number of the cells in the body are precursors to the pluripotential embryonic stems cells. Stem cells are vital to the development of an organism. Throughout the life of an individual, stem cells regenerate themselves as well as develop into cells that form the blood, lymphoid and many other systems. Thus stem cells have two functions — first they divide to form daughter stem cells so that there is a pool of pluripotential stem cells kept throughout life; second, they develop into "specialized cells" of the respective system. Stem cells not only supply the progenitor cells to these systems at the respective stages of development, but also supply them with replacement cells further in life when there is a depletion of a certain kind of cell pool. A special kind of stem cell is the pluripotential hematopoietic stem cell (HSC) from which emerge the blood cells.
The model of XCI in human blood cell development
If we consider the case of blood cells and follow it through development, the following course of events would take place (from what is known/suggested in the literature). First, at the stage in development at which XCI occurs, of the number of progenitor SC there are n0 cells that form the progenitor pool of hematopoietic SC (HSC). This size of this pool n0 has been estimated to be around 20 (or 19 — Fialkow 1973; 14-16 — Tonon et al., 1998). The pool of n0 cells grows for t0 generations until it reaches a certain number of cells — n1. The cells at this stage are no longer progenitor HSC, they are HSC.
At this point in time, the n1 HSC begin to divide and die for t1 generations, keeping a pool of HSC in the body throughout life. Thus, some of the HSC that we have at age 70 are the same cells that we had at birth. Meanwhile, some of the HSC are sampled to become differentiated blood cells. After a number of cells divisions, most of the blood cells go through several stages until they fully develop and "specialize" into particular types of blood cells.
Further on in life, if the blood cell pool becomes depleted, HSC will be recruited to develop into blood cells, and supply the blood system with more cells. Thus throughout life the pool of HSC becomes smaller and smaller.
The program
A program in Pascal was developed to model XCI in human blood cells. The purpose of the program is to obtain the expected distribution of maternally inactivated blood cells in a sample of individuals.
The program goes through the process of development according to the model described above and records the number of maternally inactivated cells at four stages during life. The program then repeats the same process for m replicates, until the desired number of replicates is finished (the number of replicates is equivalent to the number of individuals in a sample). At the end the program shows a histogram of the distribution of maternally inactivated blood cells in a number of replicates (or a sample of individuals).
This is an interactive program, where the parameters of growth (initial number of cells, number of generations and sampling events), the seed that generates random numbers, and the output file need to be entered. The output file contains five distributions of the maternally inactivated blood cells in a number of replicates at five stages in development. Each distribution is also represented by its mean and variance.
Program description
The program goes through the process of development as described in The model of XCI in human blood cells during development. There are five stages in the program:
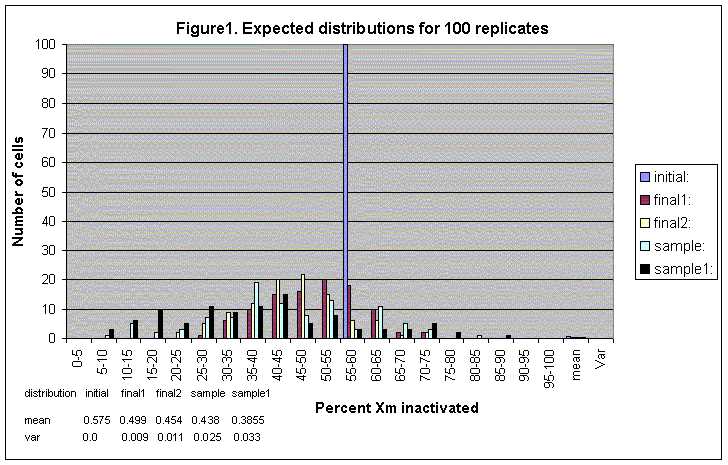
The first stage (A) is the stage of pure growth. Stage A deals with the pool of precursor HSC — of size n0, that grows for t0 generations to reach n1. In stage A, the n0 precursor cells are assigned a Xm or Xp state at random, so that if we look at the histogram of the distribution of cells Xm after a number of replicates, the graph would resemble a binomial probability distribution. The results are shown in Figure 1 (100 replicates), Figure 2 (1 000 replicates) and Figure 3 (10 000 replicates) as the series "initial". n0 and t0 have to be specified in the input. If one were to decide that the assignment of initial XCI states was not random, one would do so in the input (see Table).
After the n0 cells have each been assigned a state, they begin to grow for t0 generations. The growth is described by a division of one cell into two, one of which is itself, and the other is the daughter cells. A daughter cell always keeps the same Xp or Xm state as the mother cell. At this point in the program the cells may divide randomly, or selection may be entered — e.g. to give preferential growth to cells that are Xm. This should be specified in the input.
At stage B in development there are n1 HSC. The program counts the number of n1 cells that are Xm and records it. It does the same thing for all the replicates, and in the end it presents a frequency distribution of the cells that are Xm at this stage. The results are shown in Figure 1 (100 replicates), Figure 2 (1 000 replicates) and Figure 3 (10 000 replicates) as the series "final 1".
At this point the n1 cells begin to die and divide for t1 generations. At the same time a number of them s are being sampled at random to differentiate into cells of the blood system. Thus there is the process of birth, death and sampling occurring in this order and at constant rates. There is a constant number of cell births and deaths per each sampling event, and these parameters together with s define t1. The number of cell births and deaths per sampling event and s need to be entered in the input.
Stage C represents the pool of HSC after t1 generations of birth and death and after s events of sampling. The rates at which the HSC die and divide may be equal, or specified in the output again. Please note that if selection is chosen in the input, it acts only on the process of cell growth. The program records the number of cells that are Xm at this stage, repeats the process for all m replicates and constructs the distribution of Xm HSC cells in m individuals after a total of t0 and t1 generations have taken place. The results are shown in Figure 1 (100 replicates), Figure 2 (1 000 replicates) and Figure 3 (10 000 replicates) as the series "final 2".
Stage D represents the sample of cells that were sampled from the n1 HSC during t1. There are s cells, and for each replicate the number Xm cells is recorded. The resulting distribution represents the number of Xm cells in m individuals that are destined to become blood cells. The results are shown in Figure 1 (100 replicates), Figure 2 (1 000 replicates) and Figure 3 (10 000 replicates) as the series "sample".
After the s cells have been sampled to become blood cells, they are destined to grow (and die) until they reach the stage of fully differentiated blood cells. The s cells thus divide and die for t2 generations, until they reach a population size of s1.
Finally, stage E represents the s1 blood cells after t2 generations of births and deaths. The parameters that need to be entered in the program for this stage are the time units for t2 and the number of births and deaths per time unit at time t2 (these are all integers). Please note that if selection is entered, it acts on this stage of growth as well. After t2 the number of cells that are Xm is recorded for each of the m replicates and thus we obtain the distribution of blood cells that are maternally inactivated in m individuals. The results are shown in Figure 1 (100 replicates), Figure 2 (1 000 replicates) and Figure 3 (10 000 replicates) as the series "sample1".
Further work and uncertainties
The model of blood cell development was taken from Guilbert (Developmental Biology). I believe that the estimates of the initial number of HCS precursor pool n0 are unreliable. First, they are almost always based on XCI patterns assuming a random inactivation (or a binomial distribution). Second these estimates are often based on samples of different types of blood cells, sometimes including lymphocytes.
In terms of t0, t1 and t2, I have not been able to find realistic estimates — and therefore have chosen ‘very large numbers’ as the default values for t1 and t2. The number of sampling events s — is also unknown. Finally, if one was to compare the distribution of maternally/paternally inactivated blood cells obtained from sample of individuals to that which is expected — I believe that a 5th stage should added to the program at which a sample from the s1 blood cells is taken by the researcher to examine inactivation patterns.
Table. Possible scenarios of how skewed XCI can arise and how these can be simulated.
|
Possible cause |
Variable |
|
Clonal hematopoiesis |
Change the initial assignment of XCI states from random to that of your choice in the input. |
|
Cell depletion |
Increase the number of deaths at t1 or/and t2 |
|
Selection for/against Xm |
Introduce the relative coefficient of division |
Note:
If we have a pool of 20 n0 (precursor HSC (12 of which are Xm) that grow for 100 t0 generations and then die 80 times and divide 100 times at t1 with Xm cells having 0.8 times the relative fitness of paternally inactivated cells; then out of these HSC cells 20 s blood cells are sampled, and these blood cells divide 100 times and die 10 times during s1 in 100 m replicates; the input will be of the following form (Figure 1 is the output):
>Input the initial seed - an integer between 0 and 2147483647
236
>Input the initial number of HSC precursors n_0 (default = 20)
20
>Does XCI occur at random?(y or n)
n
>How many of the 20 HSC precursors are maternally
>inactivated?
12
>Is there selection for the growth of maternally inactivated
>cells? (y or n)'
y
>What is the relative (dis)advantage for the growth of maternally
>inactivated cells? (For growth disadvantage please enter a number
>less than 1; for growth advantage enter a number greater than 1.
>(The default value corresponding to no selection is 1)
0.8
> Input the number of generations of pure growth, t_0, for the HSC
> precursors (default = 100)
100
> Input the number of sampled precursor blood cells s (default = 20)
20
>Input the number of HSC births per sampling event, at t_1
>(default = 5)
5
>Input the number of HSC deaths per sampling event, at t_1');
>(default = 4)
4
Please enter the number of time units at t_2 (there is a constant
number of growth and death events per time unit at t_2; default = 10)
10
Please enter the number of births of blood cells per time unit at t_2
(default = 10)
10
Please enter the number of deaths of blood cells per time unit at t_2
(default = 1)
1
> Input the number of replicates m (default = 100)
100
Please enter the output file and its path (e.g. a:\results.dat)