
Lecture 25
Maintenance Elements
Introduction:
Early in Drosophila embryogenesis, transcriptional repressors encoded by the maternal-effect and segmentation genes prevent the expression of particular combinations of homeotic genes in each parasegment. During subsequent development, those homeotic genes that were initially repressed in each parasegment remain off in all the descendent cells, even though the initial repressors are absent. This phenomenon of heritable silencing depends on proteins of the Polycomb group (PcG) and on cis-acting Polycomb response elements (PREs) in the homeotic gene loci. Conversely, the trithorax group genes are required for the positive regulation of homeotic gene function and operate through cis-acting sequences known as trithorax response elements (TREs). Together, the PcG and trxG establish a form of cellular memory by faithfully maintaining transcription states determined early in embryogenesis.
It has been shown that PcG and trxG proteins colocalize at some PREs, suggesting that PcG and trxG proteins act as molecular switches through common chromosomal elements, to either direct the adjacent chromatin into an inactive or active state. A combination of genetic, molecular and biochemical approaches have provided solid evidence that PREs and TREs are intermingled, that is, closely situated but separable sequences. Accordingly, these regulatory elements that bind and respond to both groups of proteins, have been renamed 'maintenance elements' or ME.

Tillib et al., MCB 1999, 19(7): 5189-5202
Modularity:
The notion that cis-regulatory elements are composed of multiple genetic elements, or modules, was first described for viral promoters many years ago. Today, the concept that enhancers are modular is a universal concept. The reason for modularity of enhancers is that it allows transcription to be regulated in response to diverse intracellular signals. In the case of the MEs, the modularity observed most likely reflects the qualitative differences of the PcG or trxG complexes formed on each of the modules.

Taken from Bob's PhD thesis.
The diagram shown above is a summary of the biochemical and genetic analyses of the bithoraxoid 1.5kb ME. Fragments of similar colour represent modules identified in a given study. The different colours represent different studies.
The nature of Maintenance Elements:
I will be discussing the following two papers. I will not be attaching the accompanying PDFs as I will focus on the progression of the story and thus, discussing only the key experiments. If you are keen to obtain the PDFs, please let me know and I will post them.
1. Cavalli G, Paro R. The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell 1998 May 15;93(4):505-18.
2. Cavalli G, Paro R. Epigenetic inheritance of active chromatin after removal of the main transactivator. Science 1999 Oct 29;286(5441):955-8.
If time permits:
3.
Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila
enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity
that marks chromosomal Polycomb sites. Cell. 2002 Oct 18;111(2):185-96.
Paper 1.
Question: PcG and trxG proteins may act through common chromosomal elements to direct the adjacent chromatin structure into either a closed or open conformation. If so, are such chromatin states heritable throughout development in order to sustain cellular memory?

Figure 1. Description of transgenic lines used in this study.
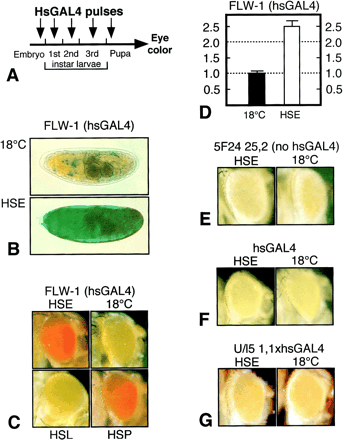
Figure 3. Release of PcG-Dependent Silencing during Embryogenesis Leads to Derepression of mini-white throughout Development
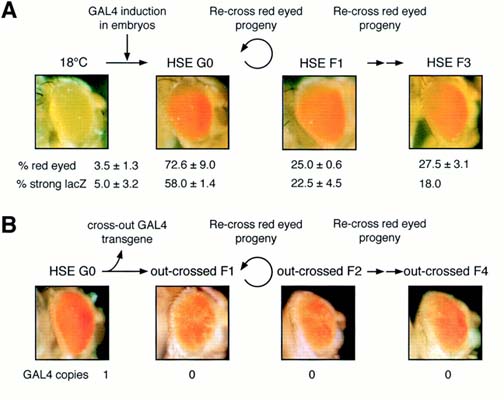
Figure 7. Meiotic Inheritance of Repressed Chromatin States.
Conclusion: Fab-7 maintenance element is a switchable chromosomal element, which can convey memory of epigenetically determined active and repressed chromatin states through mitosis and meiosis.
Paper 2.
Question: Which of the PcG and trxG proteins are involved in the mitotic and meiotic epigenetic inheritance of active and silenced chromatin states mediated by the Fab-7 ME? What are the molecular features of the chromosomal complexes involved in epigenetic inheritance through mitosis and meiosis? Does it involve histone modifications?

Figure 1. Role of PcG and trxG genes in Fab-7-mediated epigenetic inheritance.

Fig. 2. Maintenance of the active state in the presence of PcG proteins.

Fig. 3. Hyperacetylation of H4 marks epigenetically maintained open states of Fab-7.
Conclusion: Histone H4 hyperacetylation is associated with Fab-7 after activation, suggesting that H4 hyperacetylation may be a heritable epigenetic tag of the activated element.