
Methylation Levels



Aim: To review the methods used for analysing one-variable dynamical models by examining how DNA methylation levels change over time.
In vertebrates, a very large proportion of CpG sites in the genome have methylated cytosines (75-85%).
Newly replicated DNA is unmethylated. Methylases recognize sites where the parental strand is methylated and the daughter strand is not and tend to methylate the daughter strand: maintenance methylation.
In addition to maintenance methylation, occasionally a site that was not previously methylated becomes methylated: de novo methylation.
Questions:
Model: Let
 equal the proportion of methylated sites that remain methylated per cell division (maintenance methylation)
equal the proportion of methylated sites that remain methylated per cell division (maintenance methylation)
 equal the proportion of unmethylated sites that become methylated per cell division (de novo methylation)
equal the proportion of unmethylated sites that become methylated per cell division (de novo methylation)


From these definitions, we can find the proportion of methylated sites in the next cell generation:
 p[t] +
p[t] +  (1 - p[t])
(1 - p[t])The proportion of unmethylated sites in the next cell generation would be:
 ) p[t] + (1-
) p[t] + (1- ) (1 - p[t])
) (1 - p[t])Check: Is p[t+1]+q[t+1] still equal to one?


When p[t+1]=p[t], the system is at equilibrium. Let  denote this value of p.
denote this value of p.
From the recursion,  must solve
must solve
 =
= 
 +
+  (1 -
(1 -  )
)
Putting the  terms all on the same side:
terms all on the same side:
 (1-
(1-  +
+  ) =
) = 
The ONLY solution to this equation is
 =
=  /(1-
/(1-  +
+  )
)
Let f(p) be given by the recursion:
 p +
p +  (1 - p)
(1 - p)f'(p) = d(f(p))/dp is:
 -
- 
Therefore f'( ) evaluated at
) evaluated at  is (
is ( -
-  ).
).
 ) < 1 if
) < 1 if  >
>  (locally stable equilibrium)
(locally stable equilibrium)
 ) < 0 if
) < 0 if  <
<  (locally stable with an oscillatory approach to equilibrium)
(locally stable with an oscillatory approach to equilibrium)


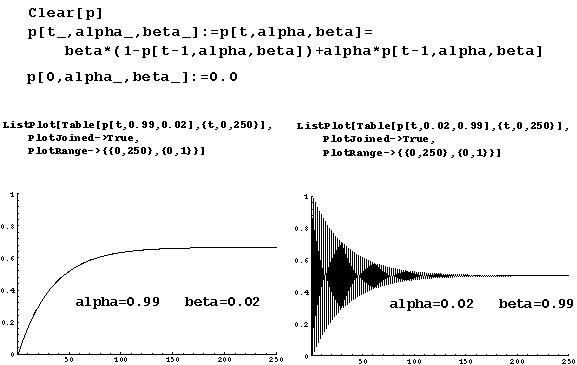
Empirical measurements indicate that maintenance methylation is about 10 to 100 times more common than de novo methylation.
Therefore, it is biologically reasonable to assume that  is greater than
is greater than  , so that a smooth approach to equilibrium is predicted.
, so that a smooth approach to equilibrium is predicted.
Finding the general solution is easier if we perform a transformation of variables.
With only one possible equilibrium, the best new variable to try is the difference between the current level of methylated CpG sites and the equilibrium level: x[t] = p[t]- .
.
The recursion then becomes
 =
=  p[t] +
p[t] +  (1 - p[t]) -
(1 - p[t]) - 
 -
-  )
)This is much easier to iterate:
 -
-  ) = x[t-2] (
) = x[t-2] ( -
-  )2...
)2...
 -
-  )t
)tThis is the general solution in terms of x[t]. In terms of p[t], we have:
 = (p[0] -
= (p[0] -  ) (
) ( -
-  )t +
)t + 


How do methylation levels change over time?
 /(1-
/(1-  +
+  ).
).
 ) (
) ( -
-  )t +
)t +  ,
,
 = (0 -
= (0 -  ) (
) ( -
-  )t +
)t + 
 (
( -
-  )t = 0.05
)t = 0.05 
 -
-  ]
] 

To proceed further, we must estimate the parameters.
In vertebrates, approximately 80% of CpG sites are methylated ( ) and
) and  is approximately 50
is approximately 50  .
.
This gives us two equations in two unknowns ( and
and  ) which we can then solve for
) which we can then solve for  and
and  :
:
 = 50
= 50 
 =
=  /(1-
/(1-  +
+  )
)
 /(1- 50
/(1- 50  +
+  )
) ) =
) = 
 = 0.0199 and
= 0.0199 and  =0.9950.
=0.9950.Therefore it takes approximately t = Log[0.05]/Log[0.9950 - 0.0199] = 118.8 cell generations for the genome to return to near equilibrium levels of methylation from a state in which no CpG sites are methylated.
Since it is estimated that 20-100 cell generations occur per individual generation for vertebrates, this indicates that methylation levels will return to near equilibrium within a few individual generations.