Fig. 7-32
After translation on ribosomes in the cytosolic compartment all proteins are processed either in the cytosol or in the ER/Golgi system.
The initial stages of protein processing involving folding.
Remember that folding of proteins takes place through interaction with chaperone proteins (see pp 139-40 and 232, 468-9).
Proteins that are not properly folded are destroyed. In the cytosol compartment they are tagged with ubiquitin and destroyed by proteasomes.
Other forms of processing of cytosolic proteins involve
In this lecture we will look at the following:
1. The Endoplasmic Reticulum
2. Exit of proteins from ER and transport to the Golgi
3. The Golgi and its dynamics
4. Processing of proteins in the ER and in the Golgi
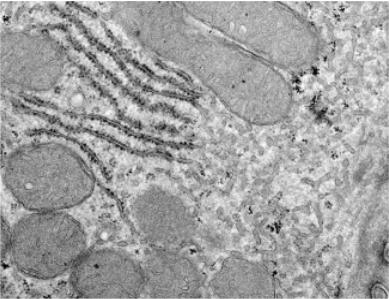
The endoplasmic reticulum (ER) consists of flattened membrane cisternae and tubules. It is continuous with the nuclear envelope and in synthetically active cells may occupy much of the cytoplasmic volume of the cell. There are two forms of ER, Rough and Smooth.
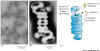
 |
Section of a liver cell showing both rough ER (left) and smooth ER (right). Click for enlarged labeled picture. |
Start by keeping in mind that proteins contained in the cisternae of the ER are separated from the cytosol by a membrane. If these proteins are to be moved, they must be moved as part of membrane vesicles, and any enzymes that act on the proteins must be contained in the vesicles or cisternae that contain the proteins.
Proteins are carried from the ER to the Golgi by vesicles (transitional vesicles). These vesicles bud from the ER cisternae through the formation of coated buds as described in the last lecture. There are two major families of coat proteins, COP and clathrin. COP proteins are involved in vesicle formation in the ER and in the cis portion of the Golgi. Clathrin, on the other hand is involved in forming vesicles in the trans Golgi network and at the cell surface.
| Make sure that you understand the use of cis and trans to indicate direction of movement relative to the ER and Golgi apparatus. See this simple animation if you have any doubt. You must understand this to make sense of what is to come. |
Control of protein exit from the ER.
Some proteins are retained in the ER (for example, the enzymes that make the oligosaccharides that are added to proteins) These proteins carry an ER retention signal (KDEL or MDEL sequence) at their carboxyl ends. See Table 14-3. Even if they get out of the ER into the cis cisternae of the Golgi, their ER targeting signal gets them sorted into vesicles that bring them back to the ER. This cis-ward movement of vesicles is called retrograde movement.
Exit of proteins from the ER is highly controlled. Proteins that are normally exported from the E R must be properly folded. Abnormally proteins are retained by chaperone molecules. Many multi-polypeptide proteins, such as antibodies, are assembled in the E R. If these proteins are not properly assembled (via formation of disulfide bridges), the proteins, like those that are not properly folded are degraded.
Cells make lots of mistakes in the assembly of proteins. They just do not let them be seen in public.
So far we have considered three large membranous organelles:
The fourth is the Golgi complex, first described at the light microscope level by Golgi (surprise!) in the 1890's in fixed material from owl brain. George Palade demonstrated the endoplasmic reticulum in 1953 by electron microscopy and shortly afterward the Golgi. These structures are hard to study without proper fixation procedures because they are very dynamic with a high turnover.
The behaviour of the Golgi depends on the presence of other organelles, e.g. cytoskeleton for support and movement.
This brief tour of the Golgi apparatus you may help you to get oriented:
Here are some images of Golgi apparatus . See if you can identify the cis and trans faces of Golgi, the trans Golgi network and transport vesicles in these pictures.
Within the Golgi there is a net flow of material (proteins) from cis to trans. This is demonstrated by labeling experiments in which cells are given labeled amino acids that are incorporated into proteins. The radioactive proteins first appear in the ER, then in the cis region of the Golgi, then in the trans region of the Golgi and finally in secretory vesicles.
Despite the flow of proteins through the Golgi apparatus, each part of the organelle has specific proteins that are resident in that region. For example, there are specific proteins that are located in the cis cisterna of the Golgi. There are enzymes that are found in the medial cisternae, and others that are located in the trans cisterna or in the trans Golgi net work. This is not a new idea. As we have already mentioned that there are proteins that are resident in the ER.
Here are some pictures showing localization of three different proteins in different regions of the Golgi apparatus:
Golgi Dynamics. How can it happen that the resident proteins appear to remain in place while the transient proteins, destined for other sites in the cell, move through the organelle in a cis to trans direction?
Over the years a number of ideas have been put forth they fall into two general models.
1. Vesicle Transport Model. This model assumes that the cisternae are essentially stationary and contain their resident proteins. The transient proteins are selected and concentrated in vesicles by the process of vesicle formation that is driven by coat proteins and their interaction with cargo receptor proteins as described in the last lecture. See vesicle formation animation for review of how this works.
These transport vesicles bud from the periphery of the Golgi cisterna as shown in the picture above, and then fuse with the appropriate target cisterna (trans to the point of origin) via the normal vesicle targeting process. In this manner a transient protein makes is way down the Golgi stack, cis to trans.
If resident Golgi proteins get into a cisterna trans to their normal location, they are presumably returned by retrograde (backward) vesicle movement in the cis direction, similar to that which occurs when resident ER proteins get carried to the Golgi.
This model is supported by your text book.
2. Cisternal Maturation Model. This model assumes that new cis cisternae are continually formed by the fusion of vesicles in the cis Golgi network. The previous cis-most cisterna is now trans to the new cis cisterna. In this model the trans cisterna is the oldest and it started life as the cis cisterna. The trans cisterna breaks up into tubules and vesicles to form the trans Golgi network.
The transient proteins stay in place and enjoy the ride. Accounting for the resident proteins is the hard part. We have to assume that the resident proteins have targeting signals that allow them to be sorted and concentrated into vesicles and shipped back to where they belong, via retrograde vesicle transport in the cis direction.
This is an older model, that despite very clear evidence from some organisms that it did occur, was discarded in favor of the vesicle model. Recently, more and more evidence is coming to light that supports the notion of cisternal maturation. Link to evidence for the cisternal maturation model.
Which model is right? As with so many issues in science, competing interpretations often both turn out to be right. It is usually a matter of the extent to which one or the other process is used in a specific example. What is very clear is that there is substantial flux of transport vesicles in the Golgi apparatus and that they move material in both cis and trans directions.
How might you distinguish between these models? This is an interesting question.
The trans Golgi Network is a major protein sorting center. Vesicles leave the trans Golgi networkfor a number of destinations. These include:
 Fig. 14-17 |
Here is a summary diagram showing the relation of the Golgi apparatus to the ER and to various target organelles, including the cell surface (plasma membrane). |
What sort of information is required for targeting?
Think of what is required for proper delivery in the case of a postal address. How specific does the information have to be. Where does the information reside? Which information is needed first? If you are uncertain about vesicle targeting, review the last lecture on vesicle transport.
Processing of proteins is initiated in the endoplasmic reticulum and continues in the Golgi apparatus.
Protein Folding and formation of disulfide bridges. Processing of proteins begins with interaction of the newly made peptide with chaparone proteins in the lumen of the ER. Chaparone proteins are released as the protein assumes its proper configuration. For many secreted proteins (including cell surface proteins), the three dimensional structure of the protein is supported by disulfide bridges that are formed between cysteine residues. It is important to keep in mind that these bonds are formed as the protein assumes its folded structure. The enzyme protein disulfide isomerase is resident in the ER and facilitates formation of disulfide bridges.
Would you expect to find disulfide bridges in the extracellular or in the cytosolic portion of membrane proteins?
Improperly folded proteins do not normally get out of the ER.
Glycosylation. Most proteins are modified in the endoplasmic reticulum by addition of polysaccharides. This process is called glycosylation. The enzymes that carry out these reactions are located in the lumen of the ER and not in the cytosol. This means that the proteins being glycosylated are bound for secretion or are membrane proteins.
There are two type of glycosylation, called N-linked and O-linked glycosylation. Your text discusses N-linked glycosylation.
In the initial step of the process a pre-fabricated oligosaccharide unit consisting of N-acetyl-glucosamine, mannose and glucose residues is transferred from a glycolipid. The glycolipid consists of the oligosaccharide chain attached to dolichol phosphate (a phospholipid with an extremely long hydrophobic chain), which serves as a membrane anchor for the oligosaccharide chain.
The oligosaccharide chain is attached to an asparagine residue through its free amino group. The asparagine residue is part of a three- amino acid target sequence consisting of asparagine, any amino acid followed by serine or threonine. The enzyme that transfers the oligosaccharide to the protein is called oligosccharide protein glycosyl transferase (this will not be on the exam).
The oligosaccharides are assembled on the cytosol side of the membrane and then flipped to the lumen (cisterna) side of the membrane by a flippase.
Subsequent to the addition of the 14 sugar oligosaccharide chain to the protein, there are a long series of modifications of the plysaccharide chain. Some of these take place in the ER, others in the cis Golgi network, other in the cis Golgi, others in medial Golgi and yet others in the trans Golgi and trans Golgi network.
Each of these steps is carried by a different enzyme that is localized in a particular region of the Golgi.
| Some details: (For illustration. Don't memorize this) The gruesome details of Golgi Glycosylation | |
| cis- Golgi | - some of the mannose residues are removed. |
| medial- Golgi | - N- acetyl-glucosamine residues are added to the remaining mannose residues |
| trans-Golgi | - glactose and N-acetyl neuraminic acid residues are added to the N-acetyl-gulcosamine residues. |
Thought question: Given this process, why might it be beneficial to have a number of Golgi cisternae rather one big vesicle?
Proteolytic cleavage of proteins as part of processing. The later stages in processing of many secreted proteins involves proteolytic cleavage of a large proprotein to produce a smaller active protein. This typically occurs in secretory granules as they move away from the trans Golgi network. These cleavages are carried out by specific endoproteases (enzymes that cleave polypeptides at sites within the chain). This sort of cleavage is very common in the case of digestive enzymes.
Why do you think that cleavage/ activation of digestive enzymes only takes place after they are inside secretory granules? What would happen if they were activated earlier?